Zinc »
PDB 6rfs-6rof »
6rie »
Zinc in PDB 6rie: Structure of Vaccinia Virus Dna-Dependent Rna Polymerase Co- Transcriptional Capping Complex
Enzymatic activity of Structure of Vaccinia Virus Dna-Dependent Rna Polymerase Co- Transcriptional Capping Complex
All present enzymatic activity of Structure of Vaccinia Virus Dna-Dependent Rna Polymerase Co- Transcriptional Capping Complex:
2.1.1.56; 2.7.7.6;
2.1.1.56; 2.7.7.6;
Other elements in 6rie:
The structure of Structure of Vaccinia Virus Dna-Dependent Rna Polymerase Co- Transcriptional Capping Complex also contains other interesting chemical elements:
| Magnesium | (Mg) | 2 atoms |
Zinc Binding Sites:
The binding sites of Zinc atom in the Structure of Vaccinia Virus Dna-Dependent Rna Polymerase Co- Transcriptional Capping Complex
(pdb code 6rie). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 4 binding sites of Zinc where determined in the Structure of Vaccinia Virus Dna-Dependent Rna Polymerase Co- Transcriptional Capping Complex, PDB code: 6rie:
Jump to Zinc binding site number: 1; 2; 3; 4;
In total 4 binding sites of Zinc where determined in the Structure of Vaccinia Virus Dna-Dependent Rna Polymerase Co- Transcriptional Capping Complex, PDB code: 6rie:
Jump to Zinc binding site number: 1; 2; 3; 4;
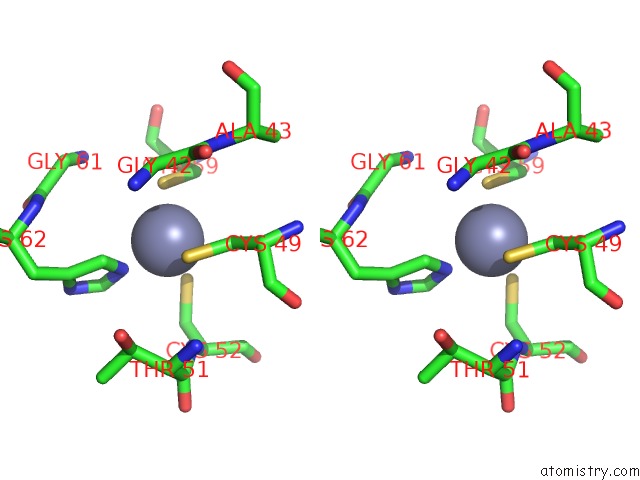
Zinc binding site 1 out of 4 in 6rie
Go back to
Zinc binding site 1 out
of 4 in the Structure of Vaccinia Virus Dna-Dependent Rna Polymerase Co- Transcriptional Capping Complex

Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Structure of Vaccinia Virus Dna-Dependent Rna Polymerase Co- Transcriptional Capping Complex within 5.0Å range:
|
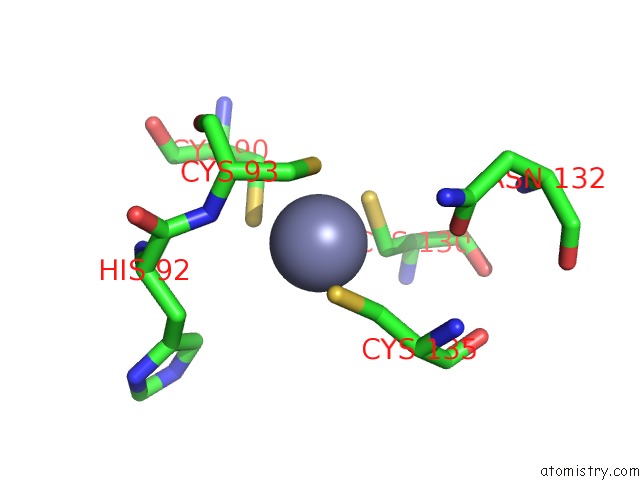
Zinc binding site 2 out of 4 in 6rie
Go back to
Zinc binding site 2 out
of 4 in the Structure of Vaccinia Virus Dna-Dependent Rna Polymerase Co- Transcriptional Capping Complex
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of Structure of Vaccinia Virus Dna-Dependent Rna Polymerase Co- Transcriptional Capping Complex within 5.0Å range:
|
Zinc binding site 3 out of 4 in 6rie
Go back to
Zinc binding site 3 out
of 4 in the Structure of Vaccinia Virus Dna-Dependent Rna Polymerase Co- Transcriptional Capping Complex

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 3 of Structure of Vaccinia Virus Dna-Dependent Rna Polymerase Co- Transcriptional Capping Complex within 5.0Å range:
|
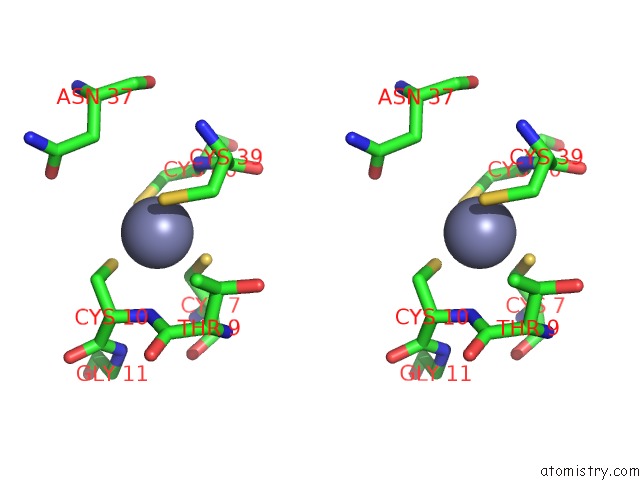
Zinc binding site 4 out of 4 in 6rie
Go back to
Zinc binding site 4 out
of 4 in the Structure of Vaccinia Virus Dna-Dependent Rna Polymerase Co- Transcriptional Capping Complex

Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 4 of Structure of Vaccinia Virus Dna-Dependent Rna Polymerase Co- Transcriptional Capping Complex within 5.0Å range:
|
Reference:
H.S.Hillen,
J.Bartuli,
C.Grimm,
C.Dienemann,
K.Bedenk,
A.A.Szalay,
U.Fischer,
P.Cramer.
Structural Basis of Poxvirus Transcription: Transcribing and Capping Vaccinia Complexes. Cell V. 179 1525 2019.
ISSN: ISSN 1097-4172
PubMed: 31835031
DOI: 10.1016/J.CELL.2019.11.023
Page generated: Tue Oct 29 06:35:03 2024
ISSN: ISSN 1097-4172
PubMed: 31835031
DOI: 10.1016/J.CELL.2019.11.023
Last articles
Zn in 9JYWZn in 9IR4
Zn in 9IR3
Zn in 9GMX
Zn in 9GMW
Zn in 9JEJ
Zn in 9ERF
Zn in 9ERE
Zn in 9EGV
Zn in 9EGW