Zinc »
PDB 4fvt-4g2w »
4g1p »
Zinc in PDB 4g1p: Structural and Mechanistic Basis of Substrate Recognition By Novel Di- Peptidase DUG1P From Saccharomyces Cerevisiae
Protein crystallography data
The structure of Structural and Mechanistic Basis of Substrate Recognition By Novel Di- Peptidase DUG1P From Saccharomyces Cerevisiae, PDB code: 4g1p
was solved by
A.K.Singh,
M.Singh,
V.K.Pandya,
V.Singh,
M.Mittal,
S.Kumaran,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 19.73 / 2.55 |
| Space group | P 65 2 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 119.132, 119.132, 176.302, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 18.6 / 23.1 |
Zinc Binding Sites:
The binding sites of Zinc atom in the Structural and Mechanistic Basis of Substrate Recognition By Novel Di- Peptidase DUG1P From Saccharomyces Cerevisiae
(pdb code 4g1p). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 2 binding sites of Zinc where determined in the Structural and Mechanistic Basis of Substrate Recognition By Novel Di- Peptidase DUG1P From Saccharomyces Cerevisiae, PDB code: 4g1p:
Jump to Zinc binding site number: 1; 2;
In total 2 binding sites of Zinc where determined in the Structural and Mechanistic Basis of Substrate Recognition By Novel Di- Peptidase DUG1P From Saccharomyces Cerevisiae, PDB code: 4g1p:
Jump to Zinc binding site number: 1; 2;
Zinc binding site 1 out of 2 in 4g1p
Go back to
Zinc binding site 1 out
of 2 in the Structural and Mechanistic Basis of Substrate Recognition By Novel Di- Peptidase DUG1P From Saccharomyces Cerevisiae

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Structural and Mechanistic Basis of Substrate Recognition By Novel Di- Peptidase DUG1P From Saccharomyces Cerevisiae within 5.0Å range:
|
Zinc binding site 2 out of 2 in 4g1p
Go back to
Zinc binding site 2 out
of 2 in the Structural and Mechanistic Basis of Substrate Recognition By Novel Di- Peptidase DUG1P From Saccharomyces Cerevisiae
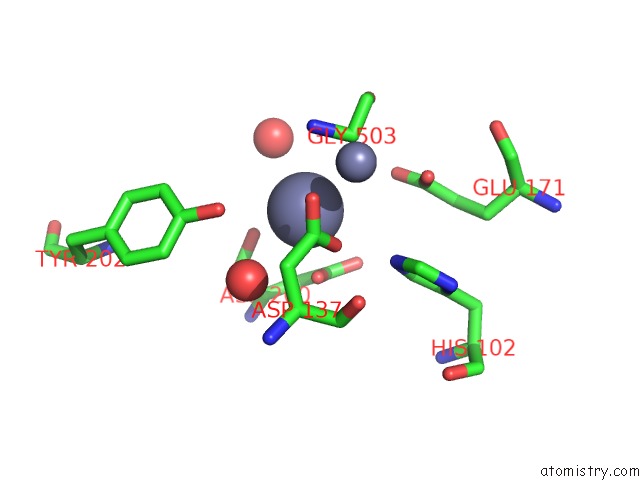
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of Structural and Mechanistic Basis of Substrate Recognition By Novel Di- Peptidase DUG1P From Saccharomyces Cerevisiae within 5.0Å range:
|
Reference:
A.K.Singh,
M.Singh,
V.K.Pandya,
V.Singh,
M.Mittal,
S.Kumaran.
Structural and Mechanistic Basis of Substrate Recognition By Novel Di-Peptidase DUG1P From Saccromyces Cerevesiae To Be Published.
Page generated: Sat Oct 26 23:05:05 2024
Last articles
Zn in 9JYWZn in 9IR4
Zn in 9IR3
Zn in 9GMX
Zn in 9GMW
Zn in 9JEJ
Zn in 9ERF
Zn in 9ERE
Zn in 9EGV
Zn in 9EGW