Zinc »
PDB 8s24-8shy »
8sez »
Zinc in PDB 8sez: Cryo-Em Structure of RYR1 + Adenine (Local Refinement of Tmd)
Zinc Binding Sites:
The binding sites of Zinc atom in the Cryo-Em Structure of RYR1 + Adenine (Local Refinement of Tmd)
(pdb code 8sez). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 4 binding sites of Zinc where determined in the Cryo-Em Structure of RYR1 + Adenine (Local Refinement of Tmd), PDB code: 8sez:
Jump to Zinc binding site number: 1; 2; 3; 4;
In total 4 binding sites of Zinc where determined in the Cryo-Em Structure of RYR1 + Adenine (Local Refinement of Tmd), PDB code: 8sez:
Jump to Zinc binding site number: 1; 2; 3; 4;
Zinc binding site 1 out of 4 in 8sez
Go back to
Zinc binding site 1 out
of 4 in the Cryo-Em Structure of RYR1 + Adenine (Local Refinement of Tmd)
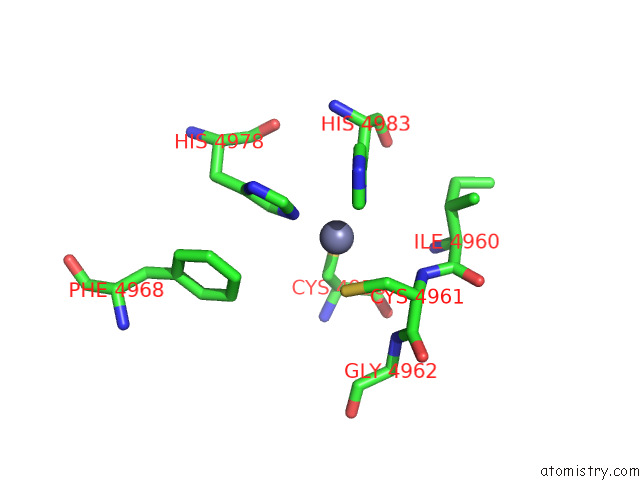
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Cryo-Em Structure of RYR1 + Adenine (Local Refinement of Tmd) within 5.0Å range:
|
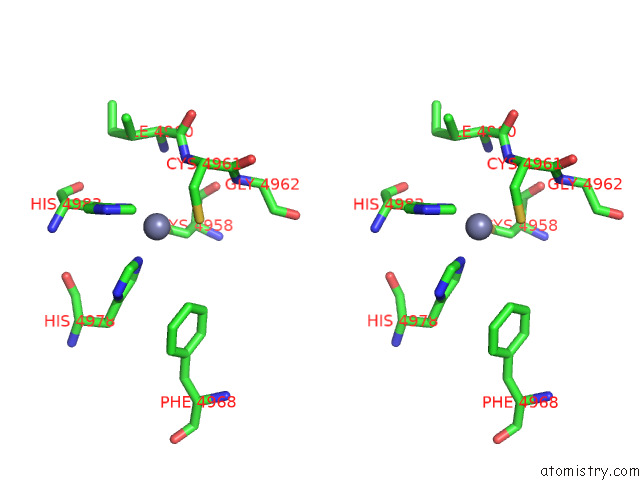
Zinc binding site 2 out of 4 in 8sez
Go back to
Zinc binding site 2 out
of 4 in the Cryo-Em Structure of RYR1 + Adenine (Local Refinement of Tmd)
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of Cryo-Em Structure of RYR1 + Adenine (Local Refinement of Tmd) within 5.0Å range:
|
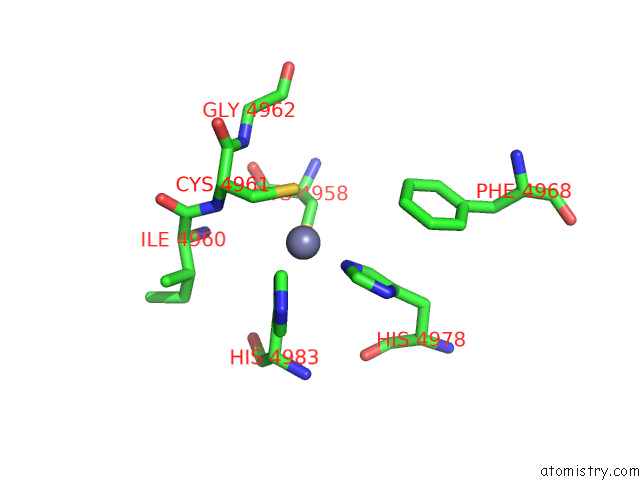
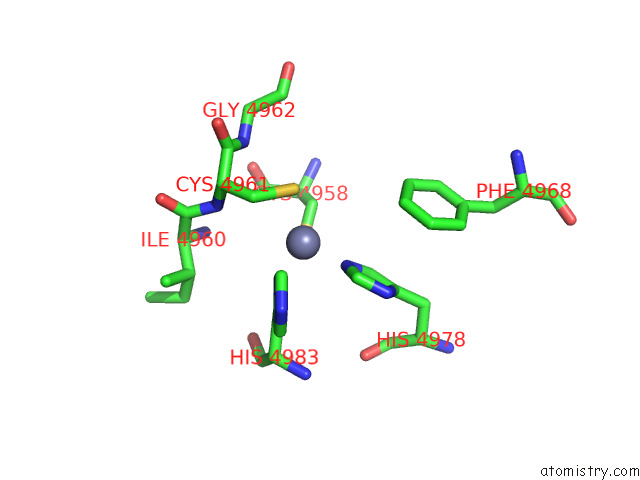
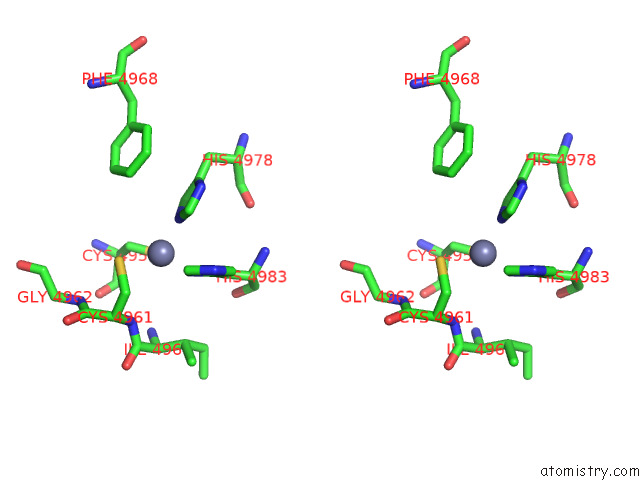
Zinc binding site 3 out of 4 in 8sez
Go back to
Zinc binding site 3 out
of 4 in the Cryo-Em Structure of RYR1 + Adenine (Local Refinement of Tmd)
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 3 of Cryo-Em Structure of RYR1 + Adenine (Local Refinement of Tmd) within 5.0Å range:
|
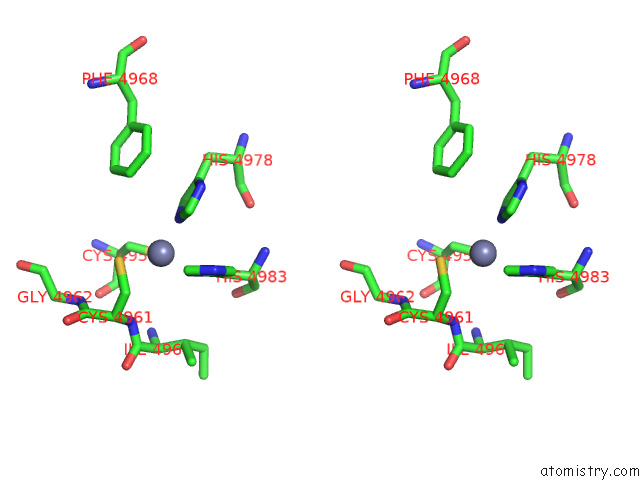
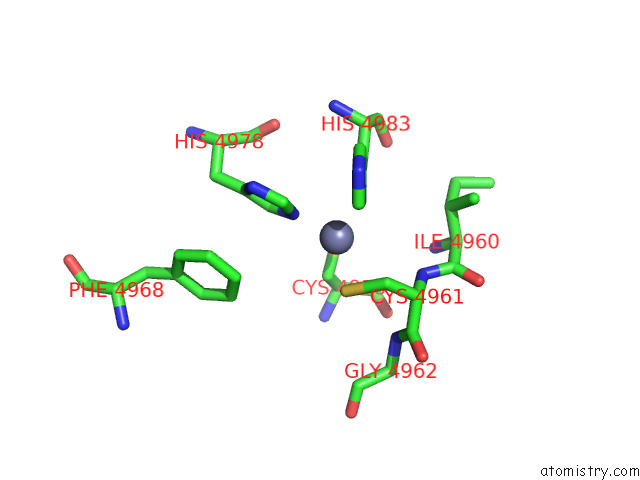
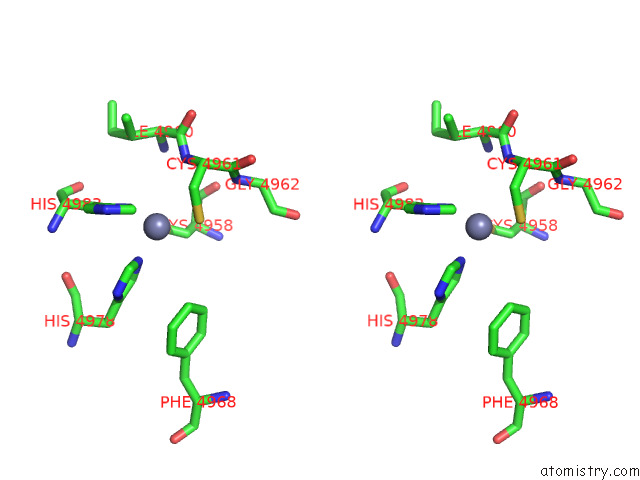
Zinc binding site 4 out of 4 in 8sez
Go back to
Zinc binding site 4 out
of 4 in the Cryo-Em Structure of RYR1 + Adenine (Local Refinement of Tmd)
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 4 of Cryo-Em Structure of RYR1 + Adenine (Local Refinement of Tmd) within 5.0Å range:
|
Reference:
S.Cholak,
J.W.Saville,
X.Zhu,
A.M.Berezuk,
K.S.Tuttle,
O.Haji-Ghassemi,
F.J.Alvarado,
F.Van Petegem,
S.Subramaniam.
Allosteric Modulation of Ryanodine Receptor RYR1 By Nucleotide Derivatives Structure 2023.
ISSN: ISSN 0969-2126
DOI: 10.1016/J.STR.2023.04.009
Page generated: Thu Oct 31 10:59:42 2024
ISSN: ISSN 0969-2126
DOI: 10.1016/J.STR.2023.04.009
Last articles
Zn in 9FD2Zn in 9GUW
Zn in 9GUX
Zn in 9F7C
Zn in 9GUR
Zn in 9F7A
Zn in 9DDE
Zn in 9DBY
Zn in 9EBZ
Zn in 9DGG