Zinc »
PDB 8s24-8shy »
8s37 »
Zinc in PDB 8s37: Dna-Bound Type IV-A3 Crispr Effector in Complex with Ding Helicase From K. Pneumoniae (State III)
Zinc Binding Sites:
The binding sites of Zinc atom in the Dna-Bound Type IV-A3 Crispr Effector in Complex with Ding Helicase From K. Pneumoniae (State III)
(pdb code 8s37). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total only one binding site of Zinc was determined in the Dna-Bound Type IV-A3 Crispr Effector in Complex with Ding Helicase From K. Pneumoniae (State III), PDB code: 8s37:
In total only one binding site of Zinc was determined in the Dna-Bound Type IV-A3 Crispr Effector in Complex with Ding Helicase From K. Pneumoniae (State III), PDB code: 8s37:
Zinc binding site 1 out of 1 in 8s37
Go back to
Zinc binding site 1 out
of 1 in the Dna-Bound Type IV-A3 Crispr Effector in Complex with Ding Helicase From K. Pneumoniae (State III)
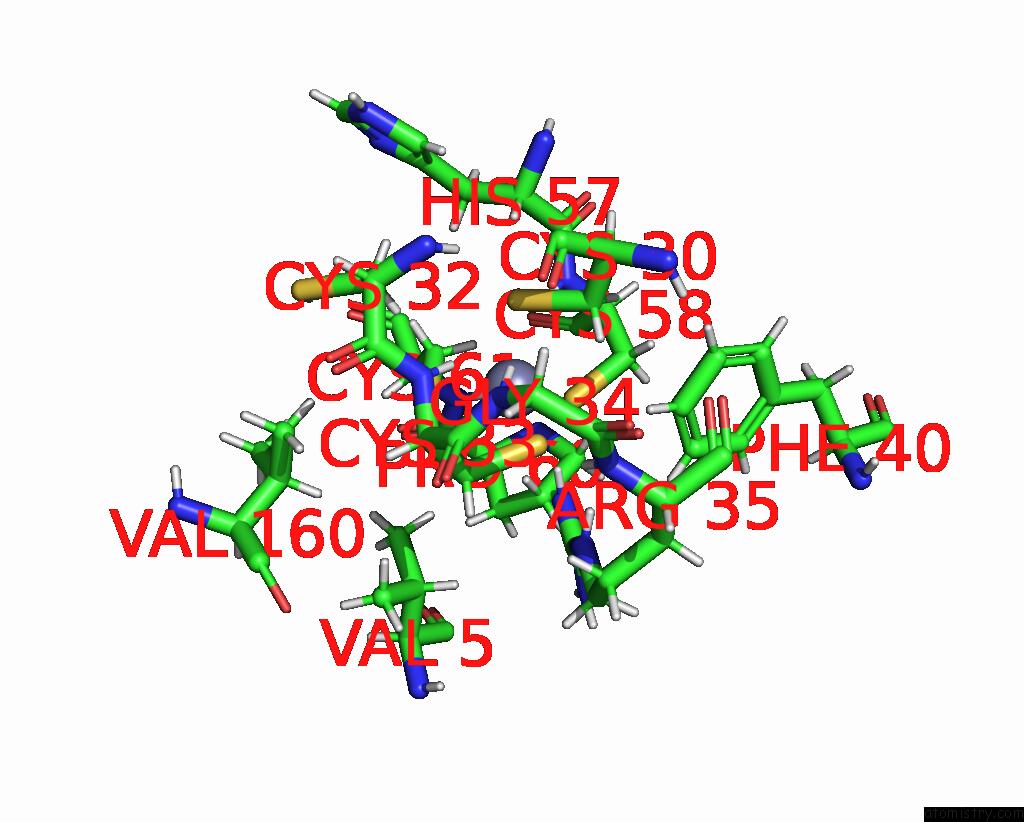
Mono view
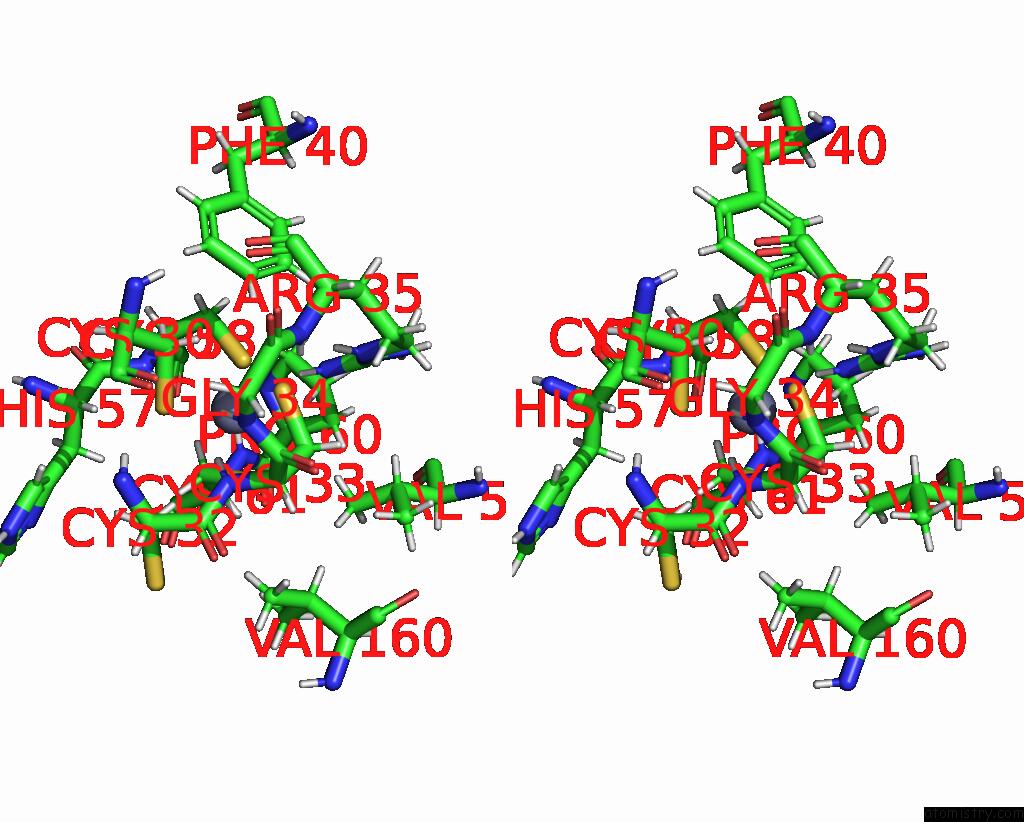
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Dna-Bound Type IV-A3 Crispr Effector in Complex with Ding Helicase From K. Pneumoniae (State III) within 5.0Å range:
|
Reference:
R.Cepaite,
N.Klein,
A.Miksys,
S.Camara-Wilpert,
V.Ragozius,
F.Benz,
A.Skorupskaite,
H.Becker,
G.Zvejyte,
N.Steube,
G.K.A.Hochberg,
L.Randau,
R.Pinilla-Redondo,
L.Malinauskaite,
P.Pausch.
Structural Variation of Types IV-A1- and IV-A3-Mediated Crispr Interference. Nat Commun V. 15 9306 2024.
ISSN: ESSN 2041-1723
PubMed: 39468082
DOI: 10.1038/S41467-024-53778-1
Page generated: Wed Nov 13 13:42:42 2024
ISSN: ESSN 2041-1723
PubMed: 39468082
DOI: 10.1038/S41467-024-53778-1
Last articles
Zn in 9FD2Zn in 9GUW
Zn in 9GUX
Zn in 9F7C
Zn in 9GUR
Zn in 9F7A
Zn in 9DDE
Zn in 9DBY
Zn in 9EBZ
Zn in 9DGG