Zinc in PDB 8ost: Structure of Human Terminal Uridylyltransferase 4 (TUT4, ZCCHC11) in Complex with Pre-LET7G Mirna and LIN28A
Enzymatic activity of Structure of Human Terminal Uridylyltransferase 4 (TUT4, ZCCHC11) in Complex with Pre-LET7G Mirna and LIN28A
All present enzymatic activity of Structure of Human Terminal Uridylyltransferase 4 (TUT4, ZCCHC11) in Complex with Pre-LET7G Mirna and LIN28A:
2.7.7.52;
2.7.7.52;
Zinc Binding Sites:
The binding sites of Zinc atom in the Structure of Human Terminal Uridylyltransferase 4 (TUT4, ZCCHC11) in Complex with Pre-LET7G Mirna and LIN28A
(pdb code 8ost). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 2 binding sites of Zinc where determined in the Structure of Human Terminal Uridylyltransferase 4 (TUT4, ZCCHC11) in Complex with Pre-LET7G Mirna and LIN28A, PDB code: 8ost:
Jump to Zinc binding site number: 1; 2;
In total 2 binding sites of Zinc where determined in the Structure of Human Terminal Uridylyltransferase 4 (TUT4, ZCCHC11) in Complex with Pre-LET7G Mirna and LIN28A, PDB code: 8ost:
Jump to Zinc binding site number: 1; 2;
Zinc binding site 1 out of 2 in 8ost
Go back to
Zinc binding site 1 out
of 2 in the Structure of Human Terminal Uridylyltransferase 4 (TUT4, ZCCHC11) in Complex with Pre-LET7G Mirna and LIN28A
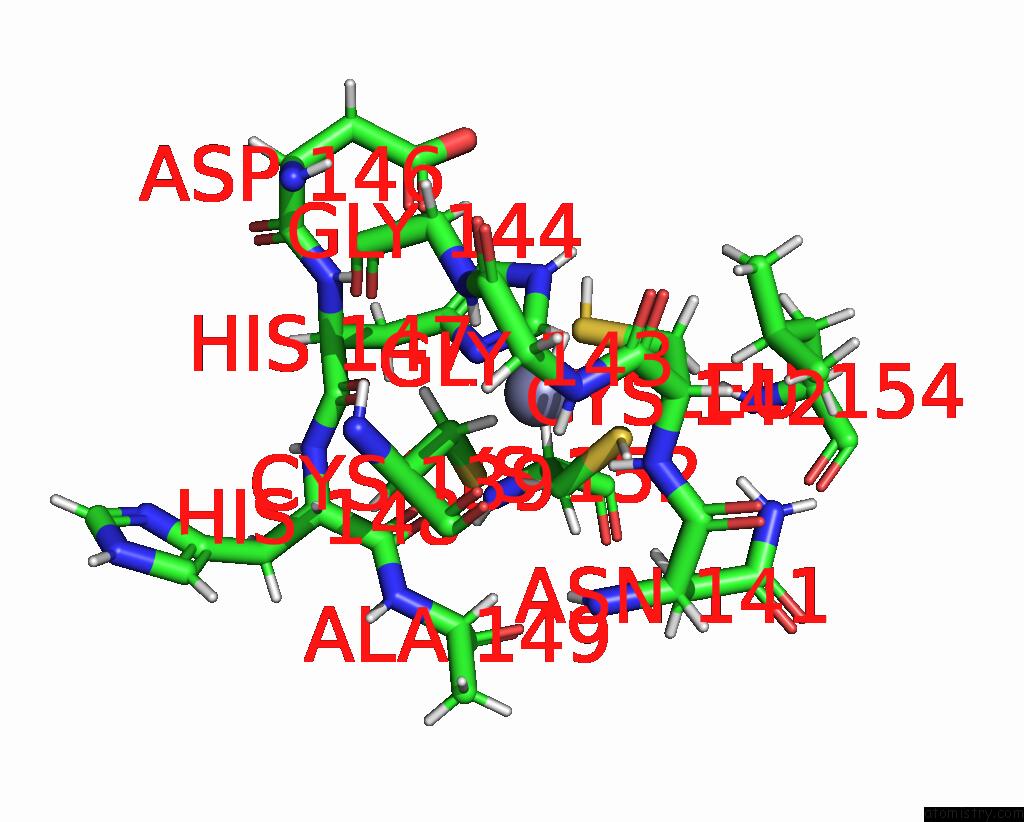
Mono view
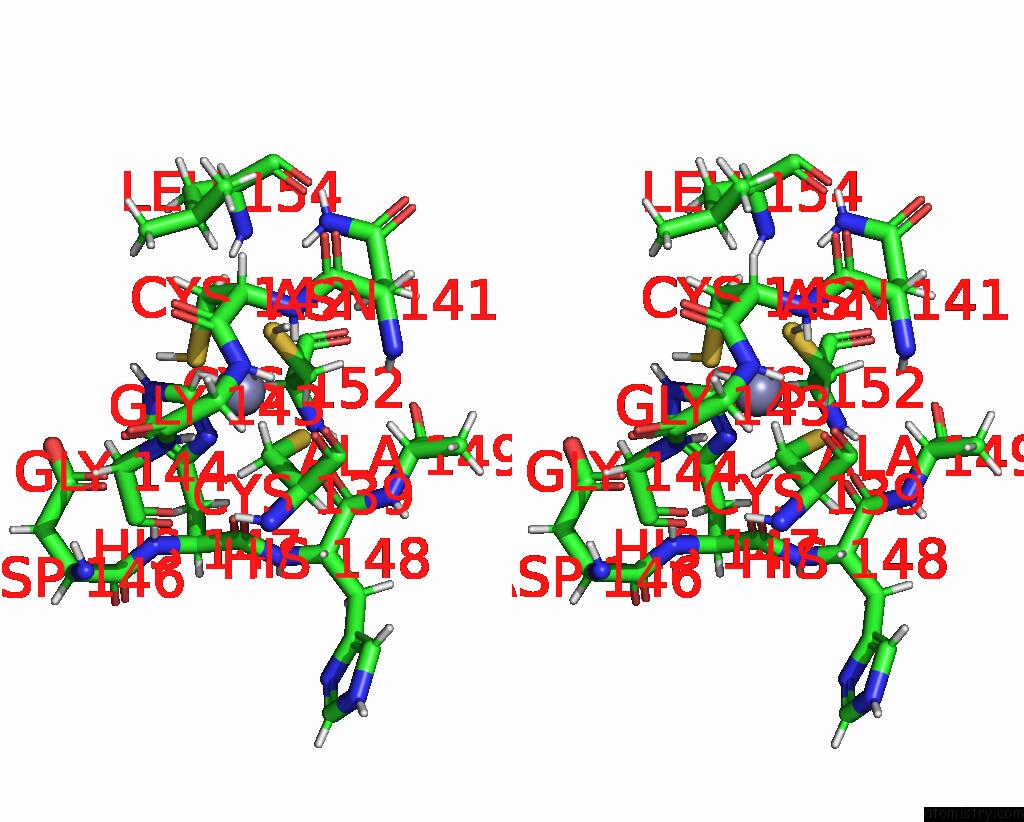
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Structure of Human Terminal Uridylyltransferase 4 (TUT4, ZCCHC11) in Complex with Pre-LET7G Mirna and LIN28A within 5.0Å range:
|
Zinc binding site 2 out of 2 in 8ost
Go back to
Zinc binding site 2 out
of 2 in the Structure of Human Terminal Uridylyltransferase 4 (TUT4, ZCCHC11) in Complex with Pre-LET7G Mirna and LIN28A
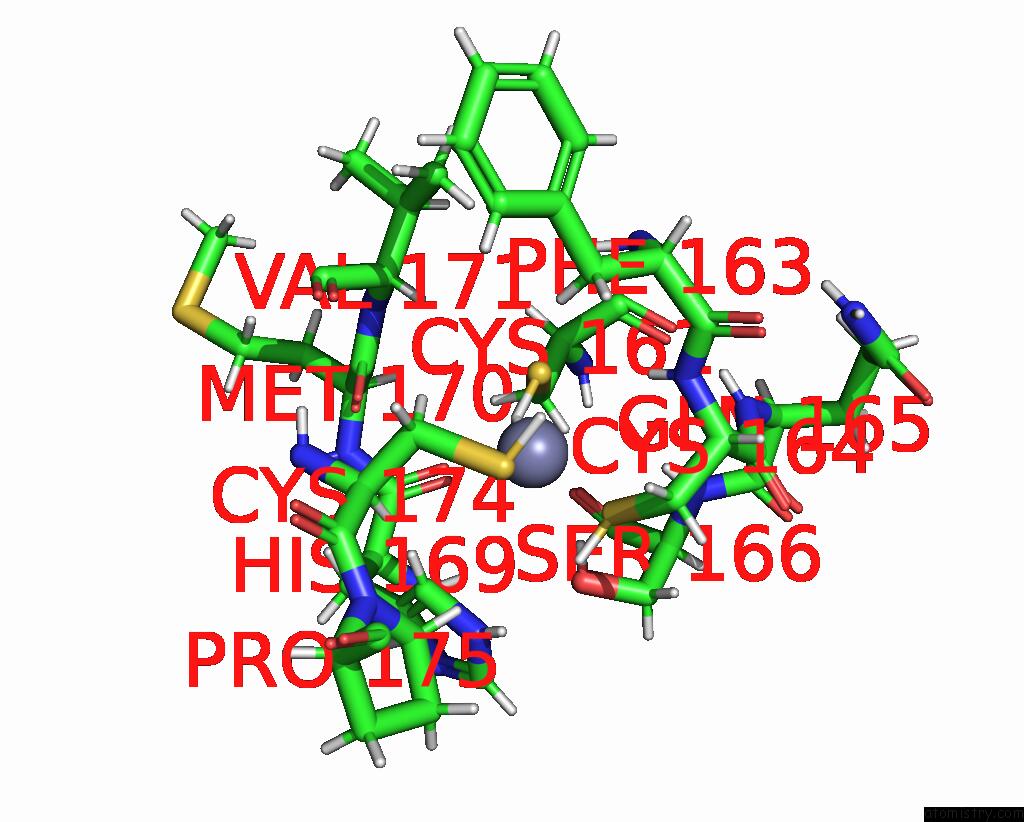
Mono view
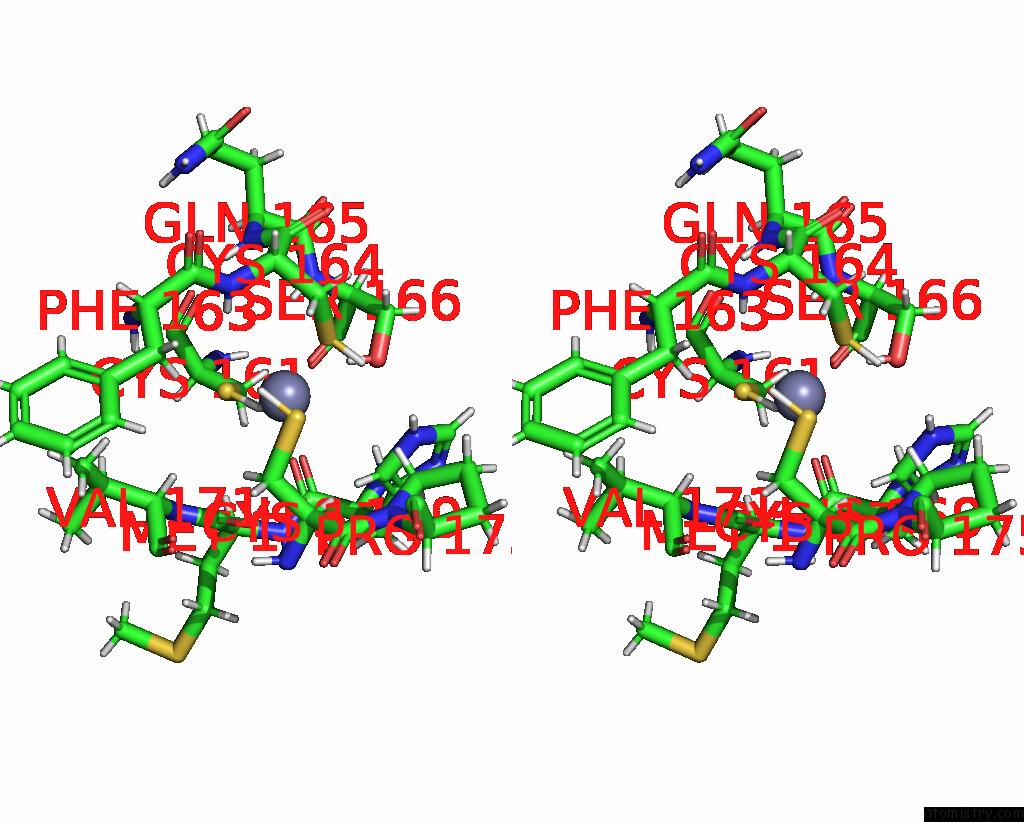
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of Structure of Human Terminal Uridylyltransferase 4 (TUT4, ZCCHC11) in Complex with Pre-LET7G Mirna and LIN28A within 5.0Å range:
|
Reference:
G.Ye,
M.Yi,
L.Carrique,
A.El-Sagheer,
T.Brown,
C.J.Norbury,
P.Zhang,
R.J.Gilbert.
Structural Basis For Activity Switching in Polymerases Determining the Fate of Let-7 Pre-Mirnas To Be Published.
Page generated: Thu Oct 31 08:56:15 2024
Last articles
Zn in 9JYWZn in 9IR4
Zn in 9IR3
Zn in 9GMX
Zn in 9GMW
Zn in 9JEJ
Zn in 9ERF
Zn in 9ERE
Zn in 9EGV
Zn in 9EGW