Zinc in PDB 8apx: Cryoem Structure of the Chikungunya Virus NSP1 Capping Pores in Covalent Complex with A 7GMP Cap Structure
Zinc Binding Sites:
Pages:
>>> Page 1 <<< Page 2, Binding sites: 11 - 12;Binding sites:
The binding sites of Zinc atom in the Cryoem Structure of the Chikungunya Virus NSP1 Capping Pores in Covalent Complex with A 7GMP Cap Structure (pdb code 8apx). This binding sites where shown within 5.0 Angstroms radius around Zinc atom.In total 12 binding sites of Zinc where determined in the Cryoem Structure of the Chikungunya Virus NSP1 Capping Pores in Covalent Complex with A 7GMP Cap Structure, PDB code: 8apx:
Jump to Zinc binding site number: 1; 2; 3; 4; 5; 6; 7; 8; 9; 10;
Zinc binding site 1 out of 12 in 8apx
Go back to
Zinc binding site 1 out
of 12 in the Cryoem Structure of the Chikungunya Virus NSP1 Capping Pores in Covalent Complex with A 7GMP Cap Structure
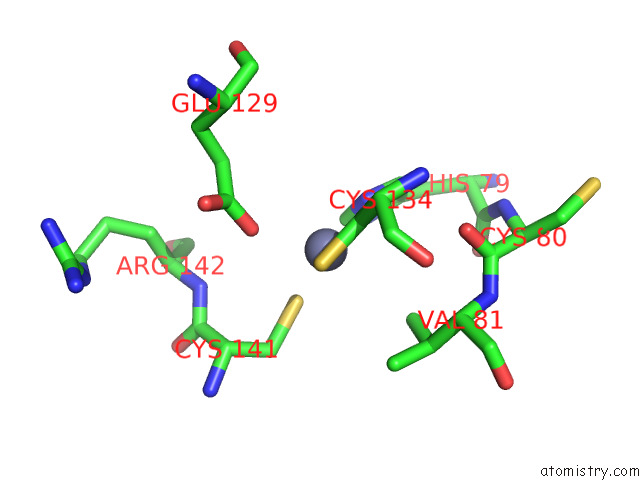
Mono view
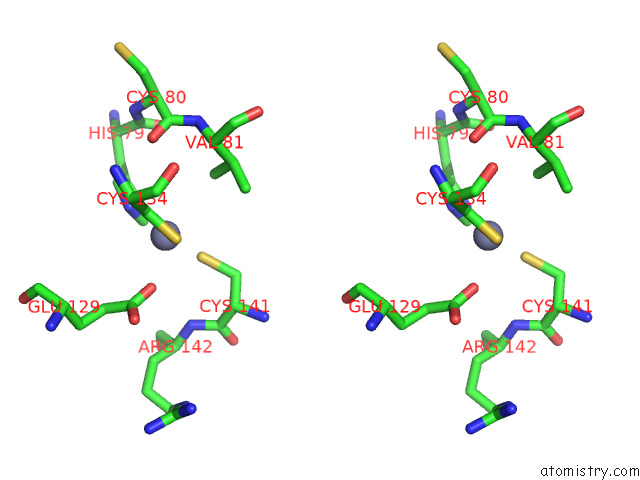
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Cryoem Structure of the Chikungunya Virus NSP1 Capping Pores in Covalent Complex with A 7GMP Cap Structure within 5.0Å range:
|
Zinc binding site 2 out of 12 in 8apx
Go back to
Zinc binding site 2 out
of 12 in the Cryoem Structure of the Chikungunya Virus NSP1 Capping Pores in Covalent Complex with A 7GMP Cap Structure
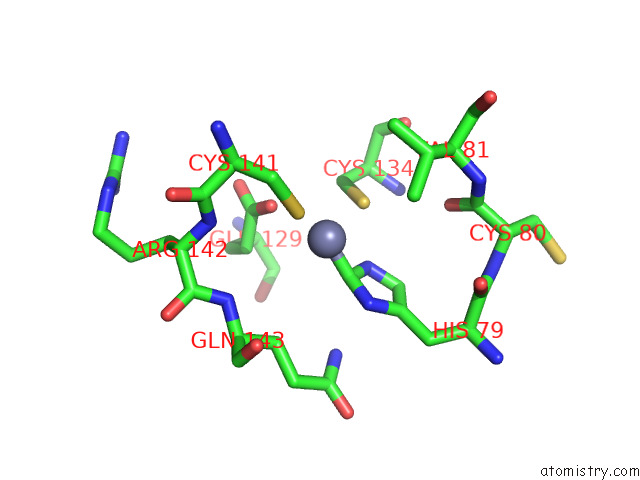
Mono view
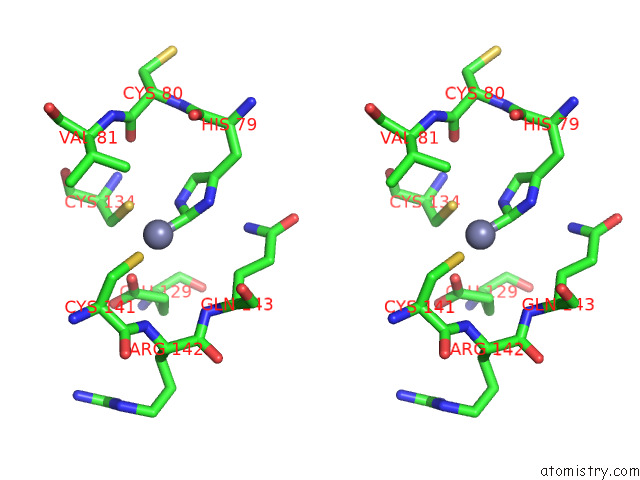
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of Cryoem Structure of the Chikungunya Virus NSP1 Capping Pores in Covalent Complex with A 7GMP Cap Structure within 5.0Å range:
|
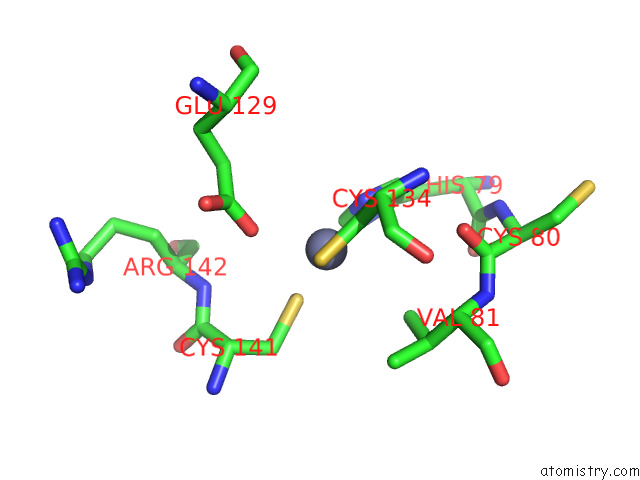
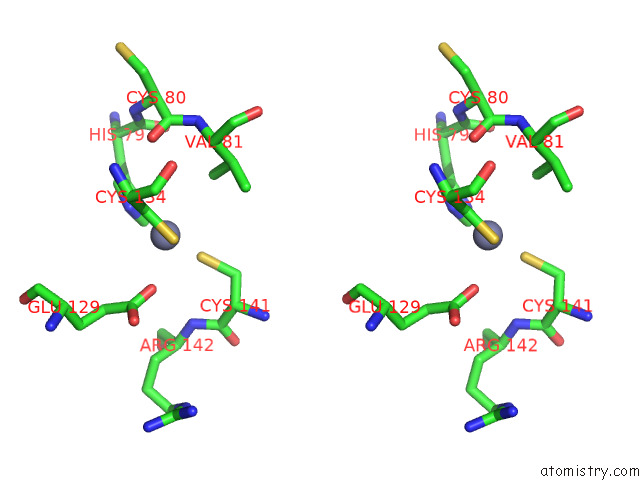
Zinc binding site 3 out of 12 in 8apx
Go back to
Zinc binding site 3 out
of 12 in the Cryoem Structure of the Chikungunya Virus NSP1 Capping Pores in Covalent Complex with A 7GMP Cap Structure
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 3 of Cryoem Structure of the Chikungunya Virus NSP1 Capping Pores in Covalent Complex with A 7GMP Cap Structure within 5.0Å range:
|
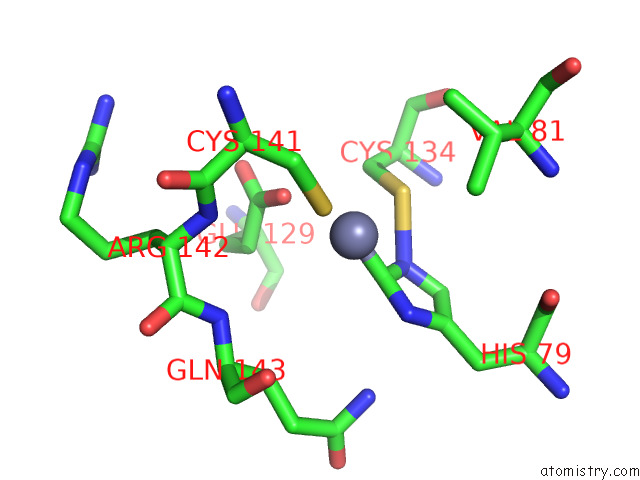
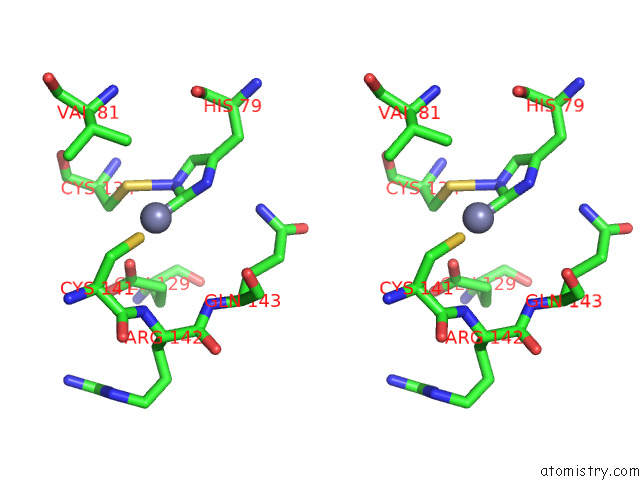
Zinc binding site 4 out of 12 in 8apx
Go back to
Zinc binding site 4 out
of 12 in the Cryoem Structure of the Chikungunya Virus NSP1 Capping Pores in Covalent Complex with A 7GMP Cap Structure
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 4 of Cryoem Structure of the Chikungunya Virus NSP1 Capping Pores in Covalent Complex with A 7GMP Cap Structure within 5.0Å range:
|
Zinc binding site 5 out of 12 in 8apx
Go back to
Zinc binding site 5 out
of 12 in the Cryoem Structure of the Chikungunya Virus NSP1 Capping Pores in Covalent Complex with A 7GMP Cap Structure
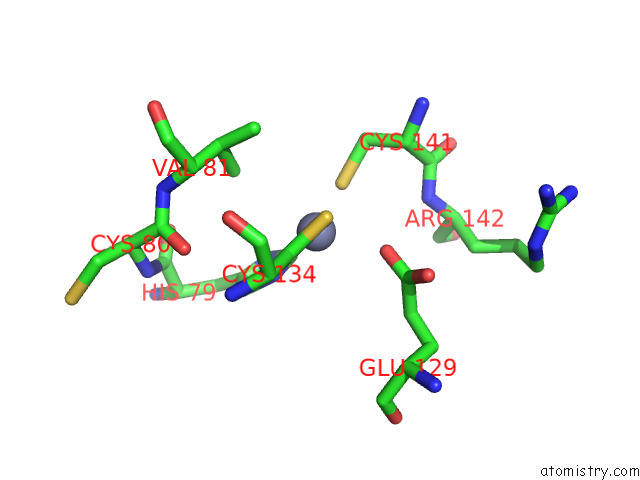
Mono view
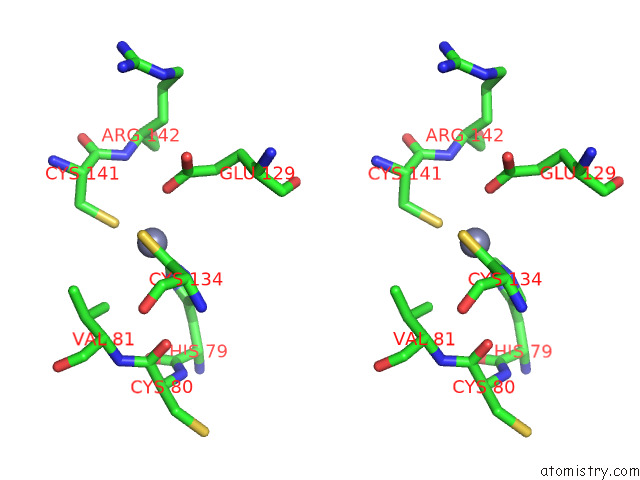
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 5 of Cryoem Structure of the Chikungunya Virus NSP1 Capping Pores in Covalent Complex with A 7GMP Cap Structure within 5.0Å range:
|
Zinc binding site 6 out of 12 in 8apx
Go back to
Zinc binding site 6 out
of 12 in the Cryoem Structure of the Chikungunya Virus NSP1 Capping Pores in Covalent Complex with A 7GMP Cap Structure
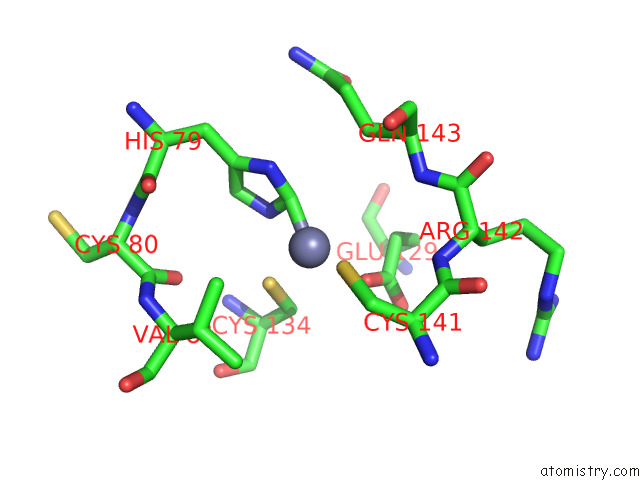
Mono view
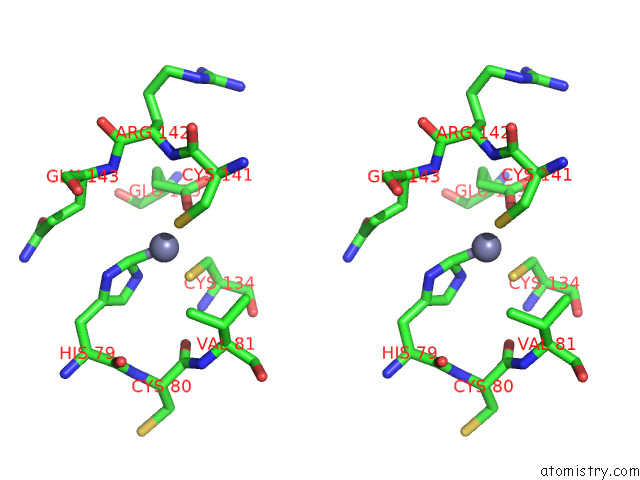
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 6 of Cryoem Structure of the Chikungunya Virus NSP1 Capping Pores in Covalent Complex with A 7GMP Cap Structure within 5.0Å range:
|
Zinc binding site 7 out of 12 in 8apx
Go back to
Zinc binding site 7 out
of 12 in the Cryoem Structure of the Chikungunya Virus NSP1 Capping Pores in Covalent Complex with A 7GMP Cap Structure
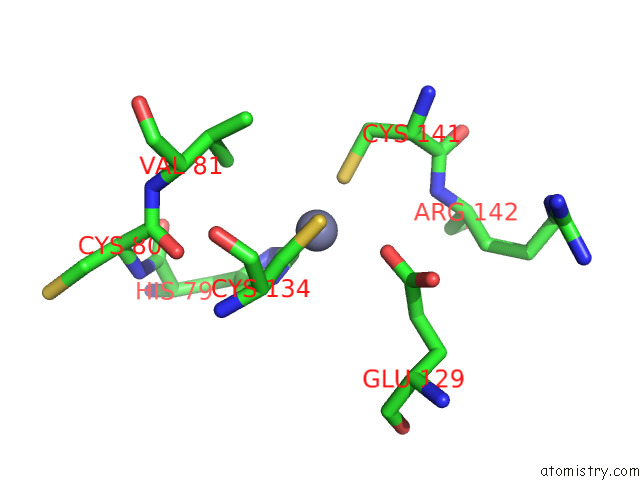
Mono view
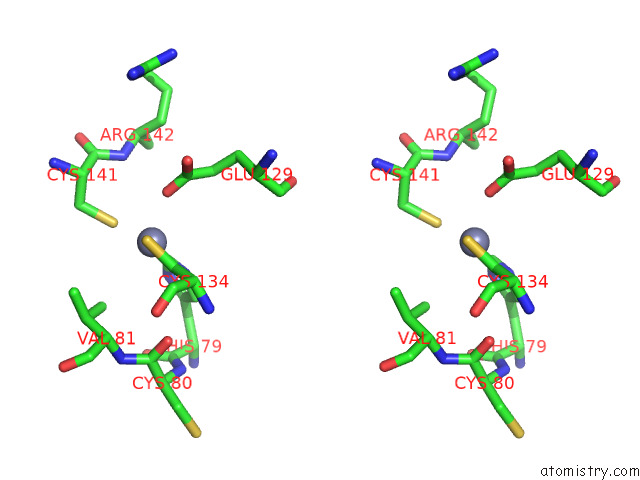
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 7 of Cryoem Structure of the Chikungunya Virus NSP1 Capping Pores in Covalent Complex with A 7GMP Cap Structure within 5.0Å range:
|
Zinc binding site 8 out of 12 in 8apx
Go back to
Zinc binding site 8 out
of 12 in the Cryoem Structure of the Chikungunya Virus NSP1 Capping Pores in Covalent Complex with A 7GMP Cap Structure
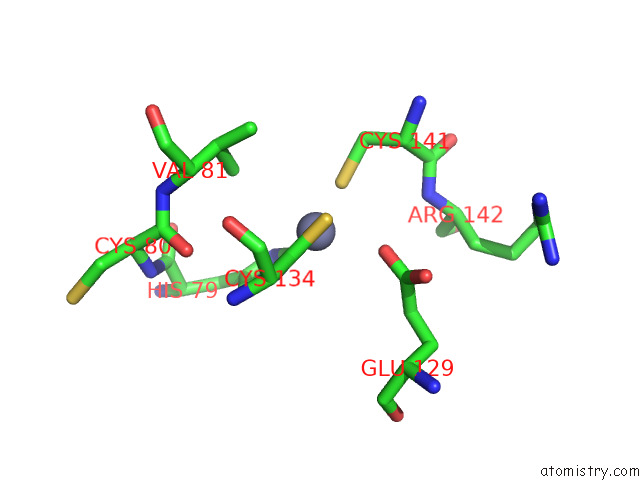
Mono view
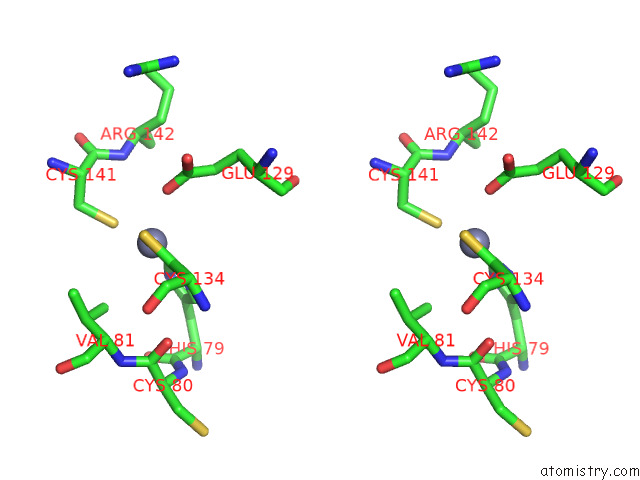
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 8 of Cryoem Structure of the Chikungunya Virus NSP1 Capping Pores in Covalent Complex with A 7GMP Cap Structure within 5.0Å range:
|
Zinc binding site 9 out of 12 in 8apx
Go back to
Zinc binding site 9 out
of 12 in the Cryoem Structure of the Chikungunya Virus NSP1 Capping Pores in Covalent Complex with A 7GMP Cap Structure
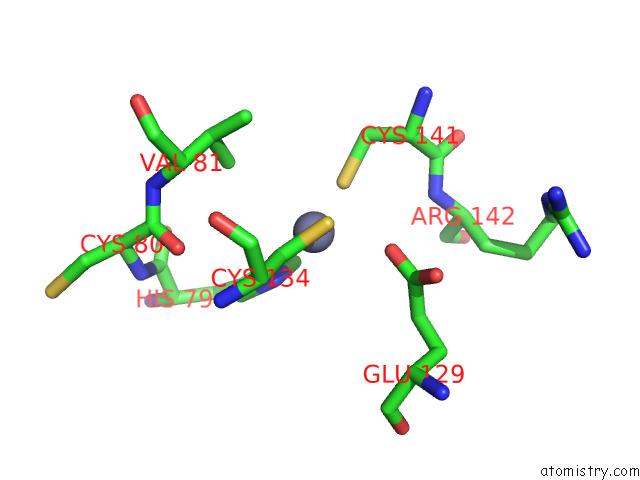
Mono view
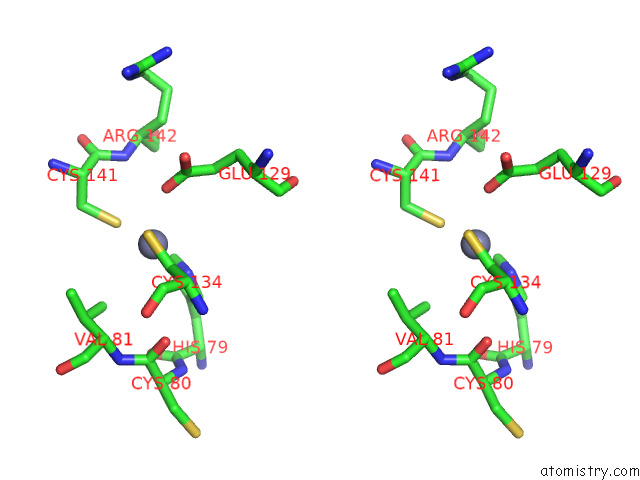
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 9 of Cryoem Structure of the Chikungunya Virus NSP1 Capping Pores in Covalent Complex with A 7GMP Cap Structure within 5.0Å range:
|
Zinc binding site 10 out of 12 in 8apx
Go back to
Zinc binding site 10 out
of 12 in the Cryoem Structure of the Chikungunya Virus NSP1 Capping Pores in Covalent Complex with A 7GMP Cap Structure
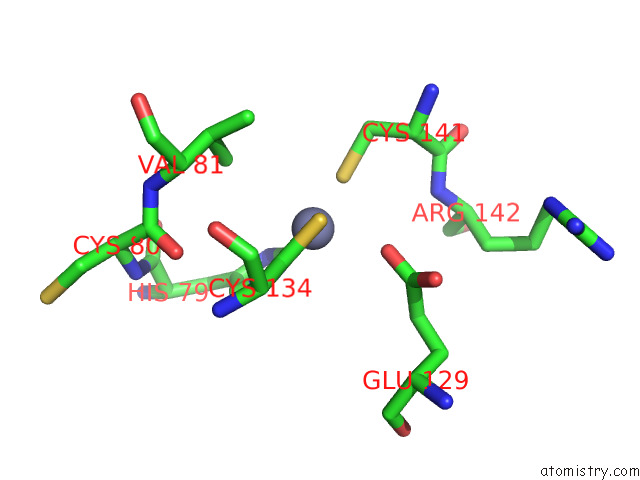
Mono view
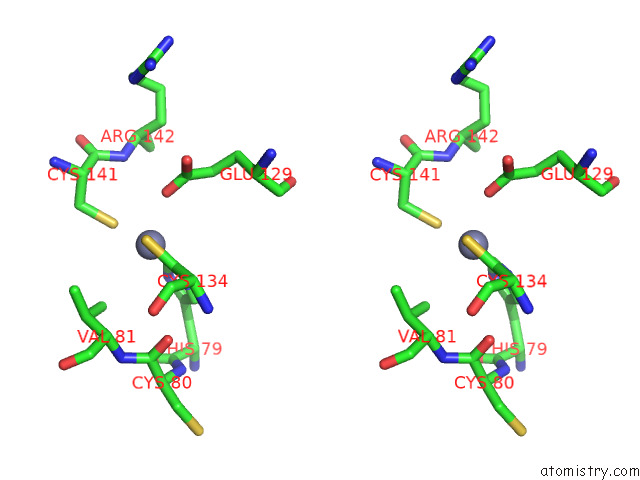
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 10 of Cryoem Structure of the Chikungunya Virus NSP1 Capping Pores in Covalent Complex with A 7GMP Cap Structure within 5.0Å range:
|
Reference:
R.Jones,
M.Hons,
N.Rabah,
N.Zamarreno,
R.Arranz,
J.Reguera.
Structural Basis and Dynamics of Chikungunya Alphavirus Rna Capping By NSP1 Capping Pores. Proc.Natl.Acad.Sci.Usa V. 120 34120 2023.
ISSN: ESSN 1091-6490
PubMed: 36913573
DOI: 10.1073/PNAS.2213934120
Page generated: Wed Oct 30 17:56:55 2024
ISSN: ESSN 1091-6490
PubMed: 36913573
DOI: 10.1073/PNAS.2213934120
Last articles
Zn in 9JYWZn in 9IR4
Zn in 9IR3
Zn in 9GMX
Zn in 9GMW
Zn in 9JEJ
Zn in 9ERF
Zn in 9ERE
Zn in 9EGV
Zn in 9EGW