Zinc »
PDB 7pcp-7pp2 »
7pok »
Zinc in PDB 7pok: Crystal Structure of Zad-Domain of Pita Protein From D.Melanogaster
Protein crystallography data
The structure of Crystal Structure of Zad-Domain of Pita Protein From D.Melanogaster, PDB code: 7pok
was solved by
K.M.Boyko,
A.N.Bonchuk,
A.Y.Nikolaeva,
P.G.Georgiev,
V.O.Popov,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 62.81 / 1.80 |
| Space group | C 2 2 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 87.265, 90.489, 105.77, 90, 90, 90 |
| R / Rfree (%) | 19 / 22.2 |
Zinc Binding Sites:
The binding sites of Zinc atom in the Crystal Structure of Zad-Domain of Pita Protein From D.Melanogaster
(pdb code 7pok). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 4 binding sites of Zinc where determined in the Crystal Structure of Zad-Domain of Pita Protein From D.Melanogaster, PDB code: 7pok:
Jump to Zinc binding site number: 1; 2; 3; 4;
In total 4 binding sites of Zinc where determined in the Crystal Structure of Zad-Domain of Pita Protein From D.Melanogaster, PDB code: 7pok:
Jump to Zinc binding site number: 1; 2; 3; 4;
Zinc binding site 1 out of 4 in 7pok
Go back to
Zinc binding site 1 out
of 4 in the Crystal Structure of Zad-Domain of Pita Protein From D.Melanogaster
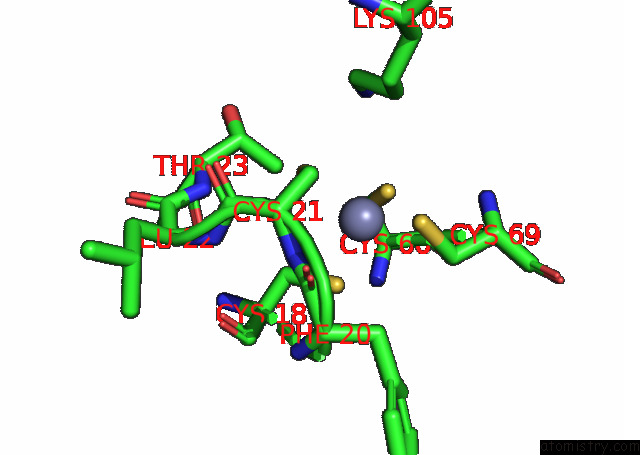
Mono view
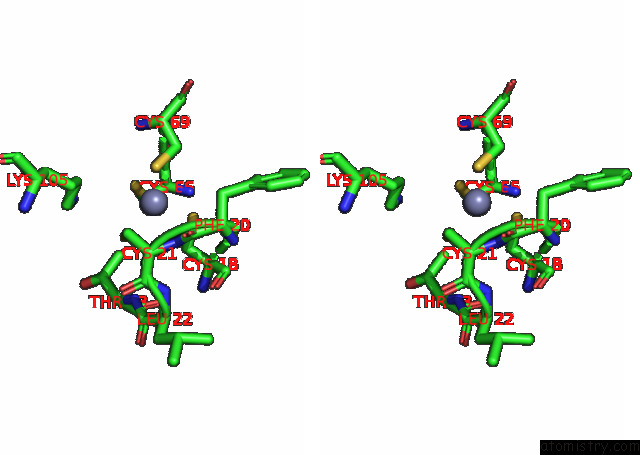
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Crystal Structure of Zad-Domain of Pita Protein From D.Melanogaster within 5.0Å range:
|
Zinc binding site 2 out of 4 in 7pok
Go back to
Zinc binding site 2 out
of 4 in the Crystal Structure of Zad-Domain of Pita Protein From D.Melanogaster

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of Crystal Structure of Zad-Domain of Pita Protein From D.Melanogaster within 5.0Å range:
|
Zinc binding site 3 out of 4 in 7pok
Go back to
Zinc binding site 3 out
of 4 in the Crystal Structure of Zad-Domain of Pita Protein From D.Melanogaster

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 3 of Crystal Structure of Zad-Domain of Pita Protein From D.Melanogaster within 5.0Å range:
|
Zinc binding site 4 out of 4 in 7pok
Go back to
Zinc binding site 4 out
of 4 in the Crystal Structure of Zad-Domain of Pita Protein From D.Melanogaster

Mono view
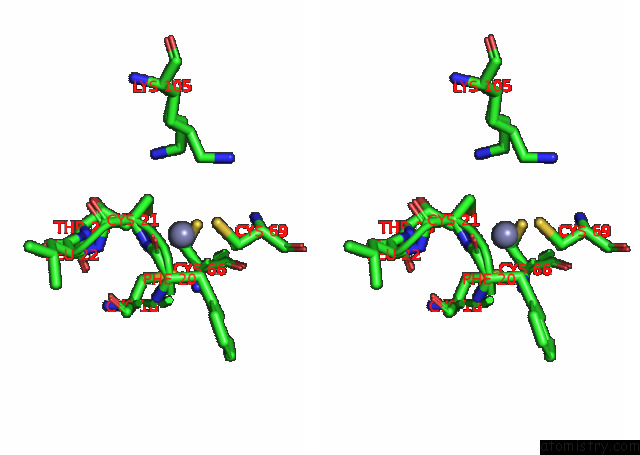
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 4 of Crystal Structure of Zad-Domain of Pita Protein From D.Melanogaster within 5.0Å range:
|
Reference:
A.N.Bonchuk,
K.M.Boyko,
A.Y.Nikolaeva,
A.D.Burtseva,
V.O.Popov,
P.G.Georgiev.
Structural Insights Into Highly Similar Spatial Organization of Zinc-Finger Associated (Zad) Domains with A Very Low Sequence Similarity To Be Published.
Page generated: Fri Aug 22 03:29:21 2025
Last articles
Zn in 8H2BZn in 8H0W
Zn in 8H0V
Zn in 8H1F
Zn in 8H1J
Zn in 8H0I
Zn in 8GZQ
Zn in 8GZR
Zn in 8H06
Zn in 8GZH