Zinc »
PDB 7ozo-7pc6 »
7p8n »
Zinc in PDB 7p8n: Tmhydabc- T. Maritima Hydrogenase with Bridge Closed
Enzymatic activity of Tmhydabc- T. Maritima Hydrogenase with Bridge Closed
All present enzymatic activity of Tmhydabc- T. Maritima Hydrogenase with Bridge Closed:
1.12.1.4;
1.12.1.4;
Other elements in 7p8n:
The structure of Tmhydabc- T. Maritima Hydrogenase with Bridge Closed also contains other interesting chemical elements:
| Iron | (Fe) | 62 atoms |
Zinc Binding Sites:
The binding sites of Zinc atom in the Tmhydabc- T. Maritima Hydrogenase with Bridge Closed
(pdb code 7p8n). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 2 binding sites of Zinc where determined in the Tmhydabc- T. Maritima Hydrogenase with Bridge Closed, PDB code: 7p8n:
Jump to Zinc binding site number: 1; 2;
In total 2 binding sites of Zinc where determined in the Tmhydabc- T. Maritima Hydrogenase with Bridge Closed, PDB code: 7p8n:
Jump to Zinc binding site number: 1; 2;
Zinc binding site 1 out of 2 in 7p8n
Go back to
Zinc binding site 1 out
of 2 in the Tmhydabc- T. Maritima Hydrogenase with Bridge Closed

Mono view
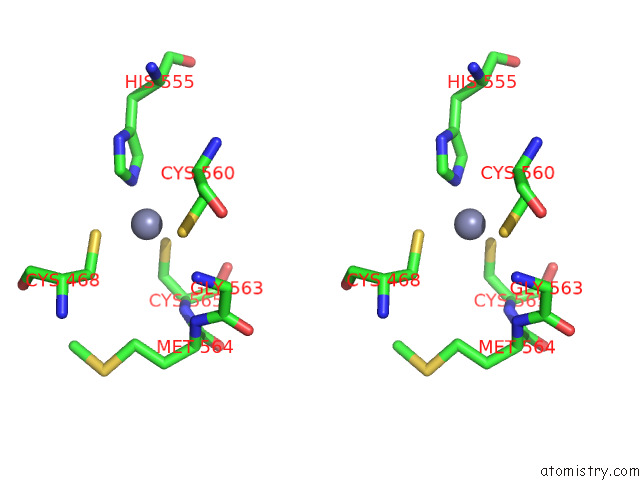
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Tmhydabc- T. Maritima Hydrogenase with Bridge Closed within 5.0Å range:
|
Zinc binding site 2 out of 2 in 7p8n
Go back to
Zinc binding site 2 out
of 2 in the Tmhydabc- T. Maritima Hydrogenase with Bridge Closed
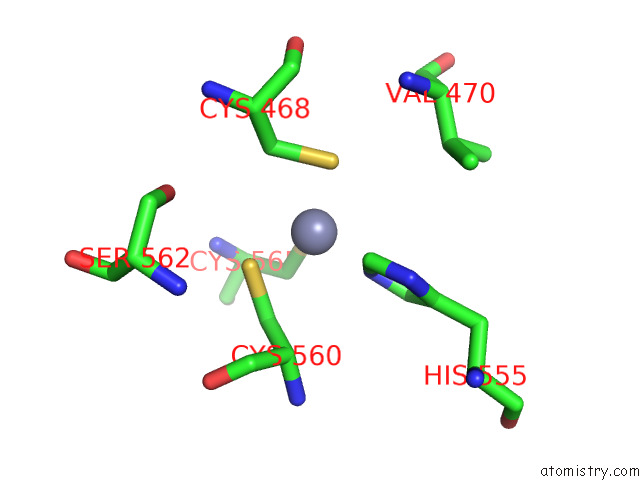
Mono view
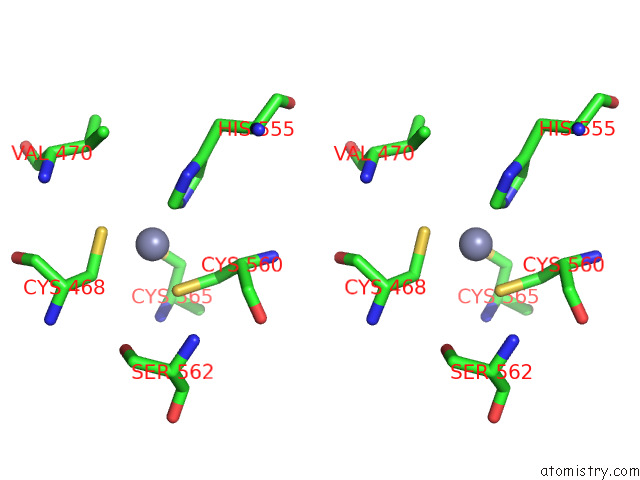
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of Tmhydabc- T. Maritima Hydrogenase with Bridge Closed within 5.0Å range:
|
Reference:
C.Furlan,
N.Chongdar,
P.Gupta,
W.Lubitz,
H.Ogata,
J.N.Blaza,
J.A.Birrell.
Structural Insight on the Mechanism of An Electron-Bifurcating [Fefe] Hydrogenase. Elife V. 11 2022.
ISSN: ESSN 2050-084X
PubMed: 36018003
DOI: 10.7554/ELIFE.79361
Page generated: Fri Aug 22 03:22:58 2025
ISSN: ESSN 2050-084X
PubMed: 36018003
DOI: 10.7554/ELIFE.79361
Last articles
Zn in 8H0VZn in 8H1F
Zn in 8H1J
Zn in 8H0I
Zn in 8GZQ
Zn in 8GZR
Zn in 8H06
Zn in 8GZH
Zn in 8GZP
Zn in 8GZC