Zinc »
PDB 5a25-5aba »
5aba »
Zinc in PDB 5aba: Structure of the P53 Cancer Mutant Y220C with Bound Small-Molecule Stabilizer PHIKAN5149
Protein crystallography data
The structure of Structure of the P53 Cancer Mutant Y220C with Bound Small-Molecule Stabilizer PHIKAN5149, PDB code: 5aba
was solved by
A.C.Joerger,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 29.44 / 1.62 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 64.893, 71.114, 104.939, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 17.8 / 20.3 |
Other elements in 5aba:
The structure of Structure of the P53 Cancer Mutant Y220C with Bound Small-Molecule Stabilizer PHIKAN5149 also contains other interesting chemical elements:
| Bromine | (Br) | 2 atoms |
| Chlorine | (Cl) | 2 atoms |
Zinc Binding Sites:
The binding sites of Zinc atom in the Structure of the P53 Cancer Mutant Y220C with Bound Small-Molecule Stabilizer PHIKAN5149
(pdb code 5aba). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 2 binding sites of Zinc where determined in the Structure of the P53 Cancer Mutant Y220C with Bound Small-Molecule Stabilizer PHIKAN5149, PDB code: 5aba:
Jump to Zinc binding site number: 1; 2;
In total 2 binding sites of Zinc where determined in the Structure of the P53 Cancer Mutant Y220C with Bound Small-Molecule Stabilizer PHIKAN5149, PDB code: 5aba:
Jump to Zinc binding site number: 1; 2;
Zinc binding site 1 out of 2 in 5aba
Go back to
Zinc binding site 1 out
of 2 in the Structure of the P53 Cancer Mutant Y220C with Bound Small-Molecule Stabilizer PHIKAN5149
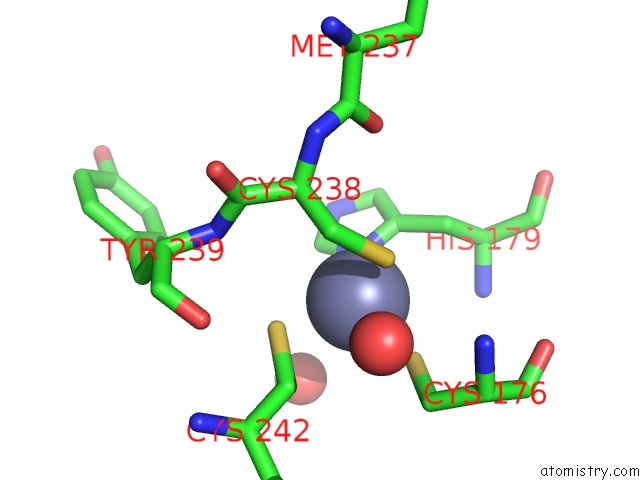
Mono view
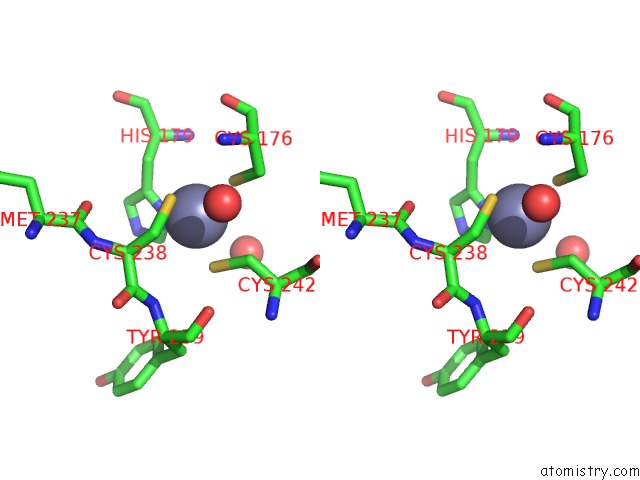
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Structure of the P53 Cancer Mutant Y220C with Bound Small-Molecule Stabilizer PHIKAN5149 within 5.0Å range:
|
Zinc binding site 2 out of 2 in 5aba
Go back to
Zinc binding site 2 out
of 2 in the Structure of the P53 Cancer Mutant Y220C with Bound Small-Molecule Stabilizer PHIKAN5149
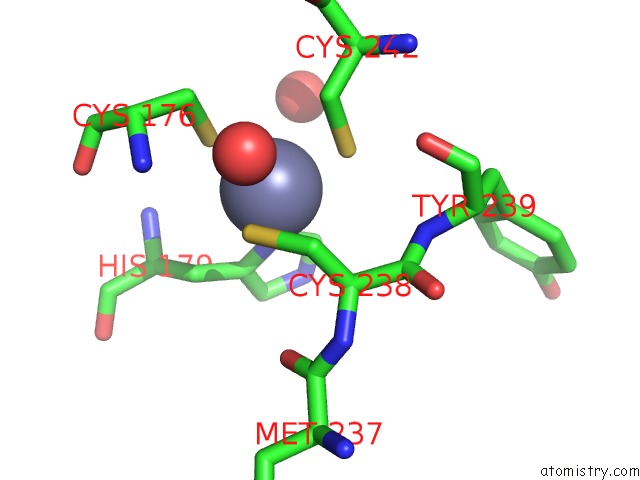
Mono view
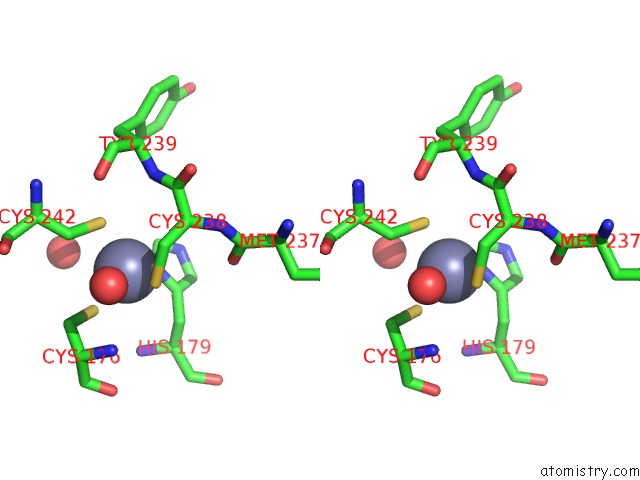
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of Structure of the P53 Cancer Mutant Y220C with Bound Small-Molecule Stabilizer PHIKAN5149 within 5.0Å range:
|
Reference:
A.C.Joerger,
M.R.Bauer,
R.Wilcken,
M.G.J.Baud,
H.Harbrecht,
T.E.Exner,
F.M.Boeckler,
J.Spencer,
A.R.Fersht.
Exploiting Transient Protein States For the Design of Small-Molecule Stabilizers of Mutant P53. Structure V. 23 2246 2015.
ISSN: ISSN 0969-2126
PubMed: 26636255
DOI: 10.1016/J.STR.2015.10.016
Page generated: Thu Aug 21 00:36:12 2025
ISSN: ISSN 0969-2126
PubMed: 26636255
DOI: 10.1016/J.STR.2015.10.016
Last articles
Zn in 6FNOZn in 6FNN
Zn in 6FN8
Zn in 6FLQ
Zn in 6FMN
Zn in 6FI9
Zn in 6FKP
Zn in 6FLP
Zn in 6FLG
Zn in 6FL8