Zinc »
PDB 1v0d-1vdd »
1va2 »
Zinc in PDB 1va2: Solution Structure of Transcription Factor SP1 Dna Binding Domain (Zinc Finger 2)
Zinc Binding Sites:
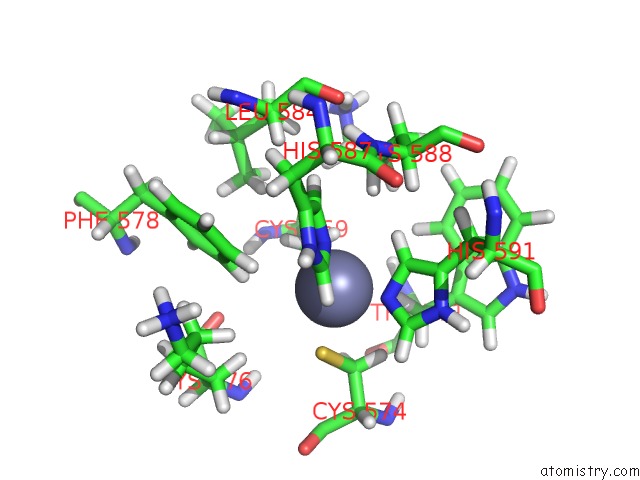
The binding sites of Zinc atom in the Solution Structure of Transcription Factor SP1 Dna Binding Domain (Zinc Finger 2)
(pdb code 1va2). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total only one binding site of Zinc was determined in the Solution Structure of Transcription Factor SP1 Dna Binding Domain (Zinc Finger 2), PDB code: 1va2:
In total only one binding site of Zinc was determined in the Solution Structure of Transcription Factor SP1 Dna Binding Domain (Zinc Finger 2), PDB code: 1va2:
Zinc binding site 1 out of 1 in 1va2
Go back to
Zinc binding site 1 out
of 1 in the Solution Structure of Transcription Factor SP1 Dna Binding Domain (Zinc Finger 2)
Mono view
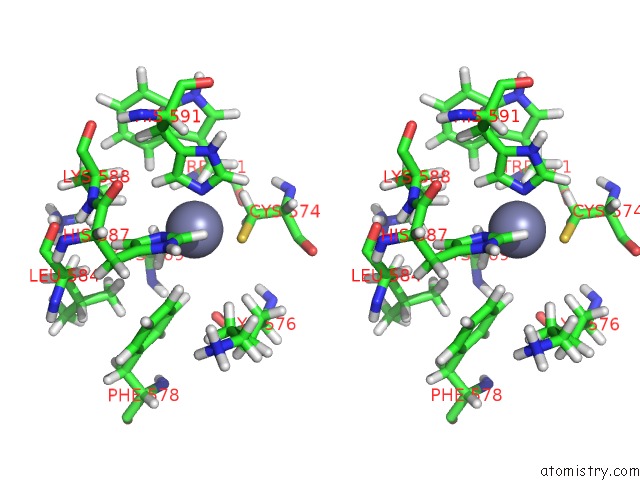
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Solution Structure of Transcription Factor SP1 Dna Binding Domain (Zinc Finger 2) within 5.0Å range:
|
Reference:
S.Oka,
Y.Shiraishi,
T.Yoshida,
T.Ohkubo,
Y.Sugiura,
Y.Kobayashi.
uc(Nmr) Structure of Transcription Factor SP1 Dna Binding Domain Biochemistry V. 43 16027 2004.
ISSN: ISSN 0006-2960
PubMed: 15609997
DOI: 10.1021/BI048438P
Page generated: Wed Oct 16 19:46:58 2024
ISSN: ISSN 0006-2960
PubMed: 15609997
DOI: 10.1021/BI048438P
Last articles
Zn in 9JPJZn in 9JP7
Zn in 9JPK
Zn in 9JPL
Zn in 9GN6
Zn in 9GN7
Zn in 9GKU
Zn in 9GKW
Zn in 9GKX
Zn in 9GL0