Zinc »
PDB 1v0d-1vdd »
1v8n »
Zinc in PDB 1v8n: Crystal Structure Analysis of the Adp-Ribose Pyrophosphatase Complexed with Zn
Enzymatic activity of Crystal Structure Analysis of the Adp-Ribose Pyrophosphatase Complexed with Zn
All present enzymatic activity of Crystal Structure Analysis of the Adp-Ribose Pyrophosphatase Complexed with Zn:
3.6.1.13;
3.6.1.13;
Protein crystallography data
The structure of Crystal Structure Analysis of the Adp-Ribose Pyrophosphatase Complexed with Zn, PDB code: 1v8n
was solved by
S.Yoshiba,
T.Ooga,
N.Nakagawa,
T.Shibata,
Y.Inoue,
S.Yokoyama,
S.Kuramitsu,
R.Masui,
Riken Structural Genomics/Proteomics Initiative (Rsgi),
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 24.93 / 1.74 |
| Space group | P 32 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 49.860, 49.860, 118.310, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 22.1 / 25.6 |
Zinc Binding Sites:
The binding sites of Zinc atom in the Crystal Structure Analysis of the Adp-Ribose Pyrophosphatase Complexed with Zn
(pdb code 1v8n). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total only one binding site of Zinc was determined in the Crystal Structure Analysis of the Adp-Ribose Pyrophosphatase Complexed with Zn, PDB code: 1v8n:
In total only one binding site of Zinc was determined in the Crystal Structure Analysis of the Adp-Ribose Pyrophosphatase Complexed with Zn, PDB code: 1v8n:
Zinc binding site 1 out of 1 in 1v8n
Go back to
Zinc binding site 1 out
of 1 in the Crystal Structure Analysis of the Adp-Ribose Pyrophosphatase Complexed with Zn
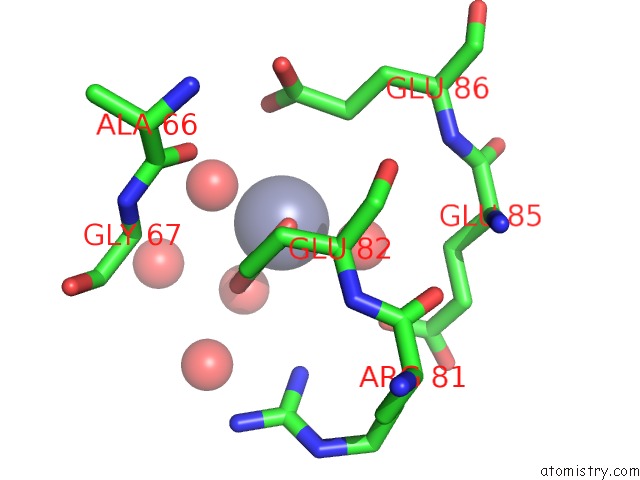
Mono view
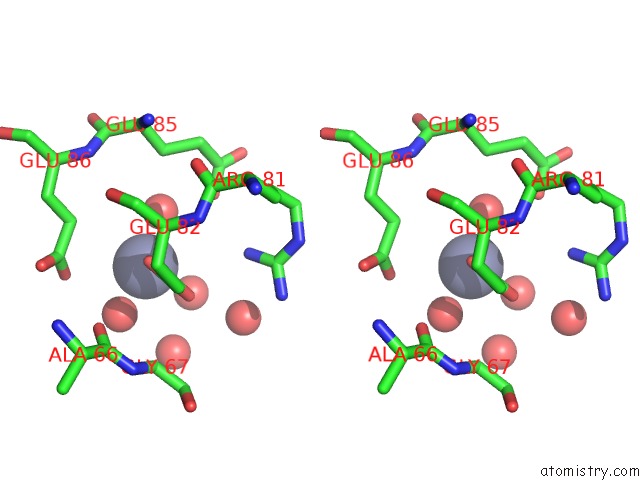
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Crystal Structure Analysis of the Adp-Ribose Pyrophosphatase Complexed with Zn within 5.0Å range:
|
Reference:
S.Yoshiba,
T.Ooga,
N.Nakagawa,
T.Shibata,
Y.Inoue,
S.Yokoyama,
S.Kuramitsu,
R.Masui.
Structural Insights Into the Thermus Thermophilus Adp-Ribose Pyrophosphatase Mechanism Via Crystal Structures with the Bound Substrate and Metal J.Biol.Chem. V. 279 37163 2004.
ISSN: ISSN 0021-9258
PubMed: 15210687
DOI: 10.1074/JBC.M403817200
Page generated: Wed Oct 16 19:45:36 2024
ISSN: ISSN 0021-9258
PubMed: 15210687
DOI: 10.1074/JBC.M403817200
Last articles
Zn in 9JPJZn in 9JP7
Zn in 9JPK
Zn in 9JPL
Zn in 9GN6
Zn in 9GN7
Zn in 9GKU
Zn in 9GKW
Zn in 9GKX
Zn in 9GL0