Zinc »
PDB 1r2y-1raf »
1r33 »
Zinc in PDB 1r33: Golgi Alpha-Mannosidase II Complex with 5-Thio-D-Mannopyranosylamine
Enzymatic activity of Golgi Alpha-Mannosidase II Complex with 5-Thio-D-Mannopyranosylamine
All present enzymatic activity of Golgi Alpha-Mannosidase II Complex with 5-Thio-D-Mannopyranosylamine:
3.2.1.114;
3.2.1.114;
Protein crystallography data
The structure of Golgi Alpha-Mannosidase II Complex with 5-Thio-D-Mannopyranosylamine, PDB code: 1r33
was solved by
D.A.Kuntz,
W.Xin,
L.M.Kavelekar,
D.R.Rose,
B.M.Pinto,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 19.86 / 1.80 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 68.838, 109.911, 138.891, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 15.6 / 18.5 |
Zinc Binding Sites:
The binding sites of Zinc atom in the Golgi Alpha-Mannosidase II Complex with 5-Thio-D-Mannopyranosylamine
(pdb code 1r33). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total only one binding site of Zinc was determined in the Golgi Alpha-Mannosidase II Complex with 5-Thio-D-Mannopyranosylamine, PDB code: 1r33:
In total only one binding site of Zinc was determined in the Golgi Alpha-Mannosidase II Complex with 5-Thio-D-Mannopyranosylamine, PDB code: 1r33:
Zinc binding site 1 out of 1 in 1r33
Go back to
Zinc binding site 1 out
of 1 in the Golgi Alpha-Mannosidase II Complex with 5-Thio-D-Mannopyranosylamine
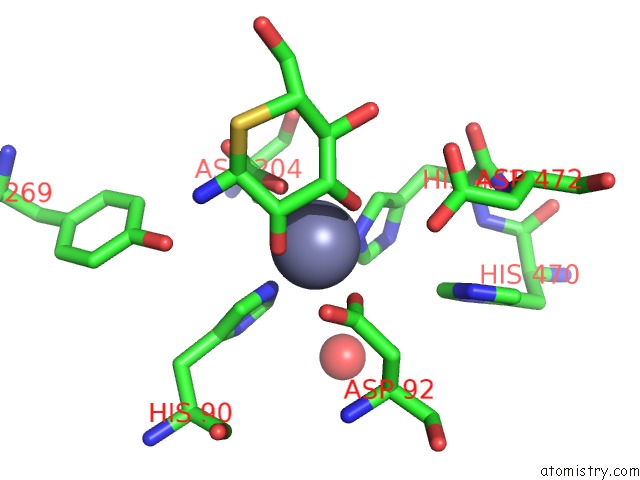
Mono view
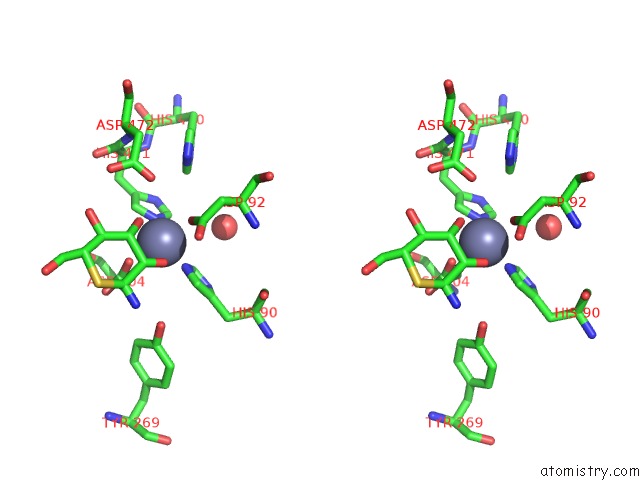
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Golgi Alpha-Mannosidase II Complex with 5-Thio-D-Mannopyranosylamine within 5.0Å range:
|
Reference:
L.M.Kavelekar,
D.A.Kuntz,
W.Xin,
B.D.Johnston,
B.Svensson,
D.R.Rose,
B.M.Pinto.
5-Thio-D-Glycopyranosylamines and Their Amidinium Salts As Potential Transition-State Mimics of Glycosyl Hydrolases: Synthesis, Enzyme Inhibitory Activities, X-Ray Crystallography, and Molecular Modeling Tetrahedron Asymmetry V. 16 1035 2005.
ISSN: ISSN 0957-4166
DOI: 10.1016/J.TETASY.2005.01.021
Page generated: Tue Aug 19 22:45:56 2025
ISSN: ISSN 0957-4166
DOI: 10.1016/J.TETASY.2005.01.021
Last articles
Zn in 1ZVLZn in 1ZVI
Zn in 1ZU1
Zn in 1ZV8
Zn in 1ZUD
Zn in 1ZT2
Zn in 1ZSC
Zn in 1ZSW
Zn in 1ZS0
Zn in 1ZR6