Zinc »
PDB 1zsc-2a2i »
1zv8 »
Zinc in PDB 1zv8: A Structure-Based Mechanism of Sars Virus Membrane Fusion
Protein crystallography data
The structure of A Structure-Based Mechanism of Sars Virus Membrane Fusion, PDB code: 1zv8
was solved by
Y.Deng,
J.Liu,
Q.Zheng,
W.Yong,
J.Dai,
M.Lu,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 70.71 / 1.94 |
| Space group | P 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 31.054, 55.565, 71.678, 81.26, 87.08, 84.10 |
| R / Rfree (%) | 20.3 / 27.4 |
Other elements in 1zv8:
The structure of A Structure-Based Mechanism of Sars Virus Membrane Fusion also contains other interesting chemical elements:
| Arsenic | (As) | 1 atom |
| Sodium | (Na) | 6 atoms |
Zinc Binding Sites:
The binding sites of Zinc atom in the A Structure-Based Mechanism of Sars Virus Membrane Fusion
(pdb code 1zv8). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 2 binding sites of Zinc where determined in the A Structure-Based Mechanism of Sars Virus Membrane Fusion, PDB code: 1zv8:
Jump to Zinc binding site number: 1; 2;
In total 2 binding sites of Zinc where determined in the A Structure-Based Mechanism of Sars Virus Membrane Fusion, PDB code: 1zv8:
Jump to Zinc binding site number: 1; 2;
Zinc binding site 1 out of 2 in 1zv8
Go back to
Zinc binding site 1 out
of 2 in the A Structure-Based Mechanism of Sars Virus Membrane Fusion
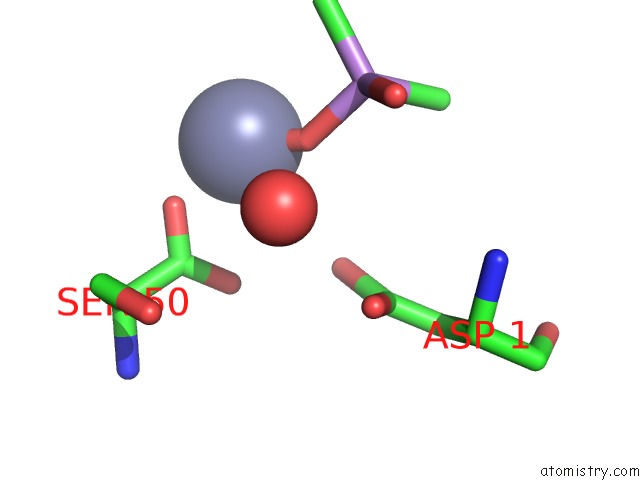
Mono view
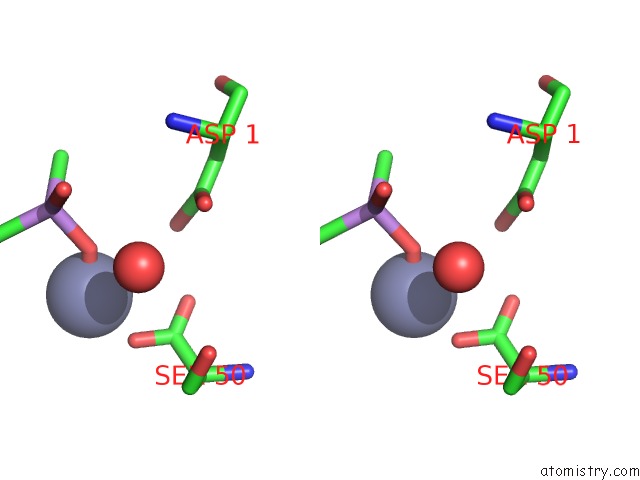
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of A Structure-Based Mechanism of Sars Virus Membrane Fusion within 5.0Å range:
|
Zinc binding site 2 out of 2 in 1zv8
Go back to
Zinc binding site 2 out
of 2 in the A Structure-Based Mechanism of Sars Virus Membrane Fusion
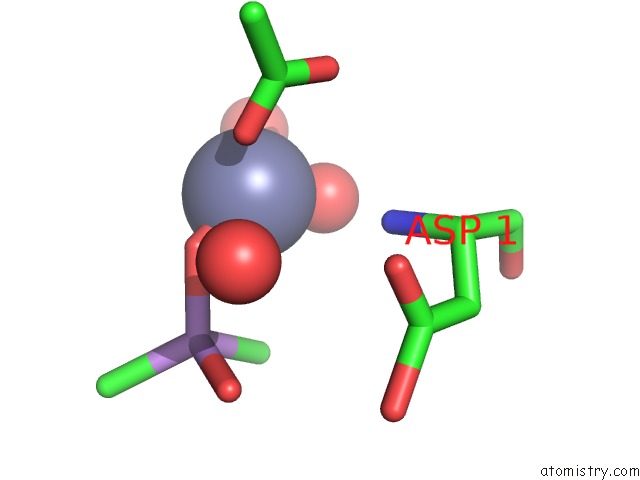
Mono view
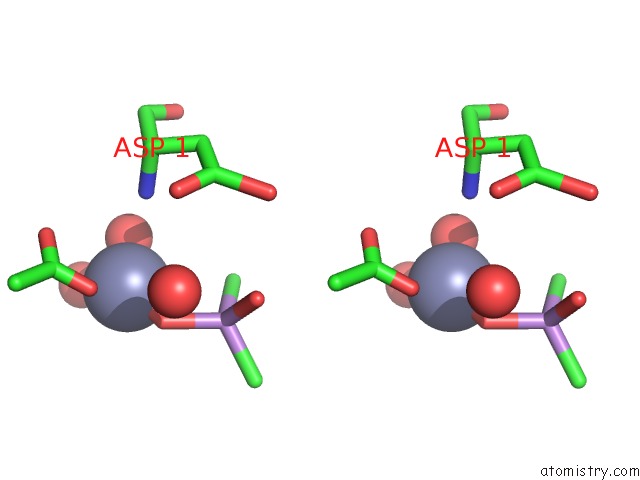
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of A Structure-Based Mechanism of Sars Virus Membrane Fusion within 5.0Å range:
|
Reference:
Y.Deng,
J.Liu,
Q.Zheng,
W.Yong,
M.Lu.
Structures and Polymorphic Interactions of Two Heptad-Repeat Regions of the Sars Virus S2 Protein. Structure V. 14 889 2006.
ISSN: ISSN 0969-2126
PubMed: 16698550
DOI: 10.1016/J.STR.2006.03.007
Page generated: Wed Aug 20 00:59:04 2025
ISSN: ISSN 0969-2126
PubMed: 16698550
DOI: 10.1016/J.STR.2006.03.007
Last articles
Zn in 2NVUZn in 2NUT
Zn in 2NUP
Zn in 2NUV
Zn in 2NSO
Zn in 2NSF
Zn in 2NR6
Zn in 2NSE
Zn in 2NQJ
Zn in 2NNX