Zinc »
PDB 2naa-2nwy »
2nso »
Zinc in PDB 2nso: Trna-Gunanine-Transglycosylase (Tgt) Mutant Y106F, C158V, A232S, V233G- Apo-Structure
Enzymatic activity of Trna-Gunanine-Transglycosylase (Tgt) Mutant Y106F, C158V, A232S, V233G- Apo-Structure
All present enzymatic activity of Trna-Gunanine-Transglycosylase (Tgt) Mutant Y106F, C158V, A232S, V233G- Apo-Structure:
2.4.2.29;
2.4.2.29;
Protein crystallography data
The structure of Trna-Gunanine-Transglycosylase (Tgt) Mutant Y106F, C158V, A232S, V233G- Apo-Structure, PDB code: 2nso
was solved by
N.Tidten,
A.Heine,
K.Reuter,
G.Klebe,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 28.42 / 1.60 |
| Space group | C 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 90.730, 64.830, 70.810, 90.00, 96.31, 90.00 |
| R / Rfree (%) | 16.2 / 22.1 |
Zinc Binding Sites:
The binding sites of Zinc atom in the Trna-Gunanine-Transglycosylase (Tgt) Mutant Y106F, C158V, A232S, V233G- Apo-Structure
(pdb code 2nso). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total only one binding site of Zinc was determined in the Trna-Gunanine-Transglycosylase (Tgt) Mutant Y106F, C158V, A232S, V233G- Apo-Structure, PDB code: 2nso:
In total only one binding site of Zinc was determined in the Trna-Gunanine-Transglycosylase (Tgt) Mutant Y106F, C158V, A232S, V233G- Apo-Structure, PDB code: 2nso:
Zinc binding site 1 out of 1 in 2nso
Go back to
Zinc binding site 1 out
of 1 in the Trna-Gunanine-Transglycosylase (Tgt) Mutant Y106F, C158V, A232S, V233G- Apo-Structure
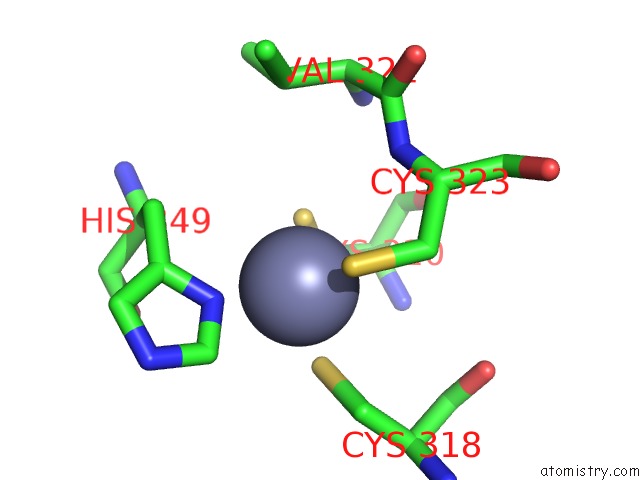
Mono view
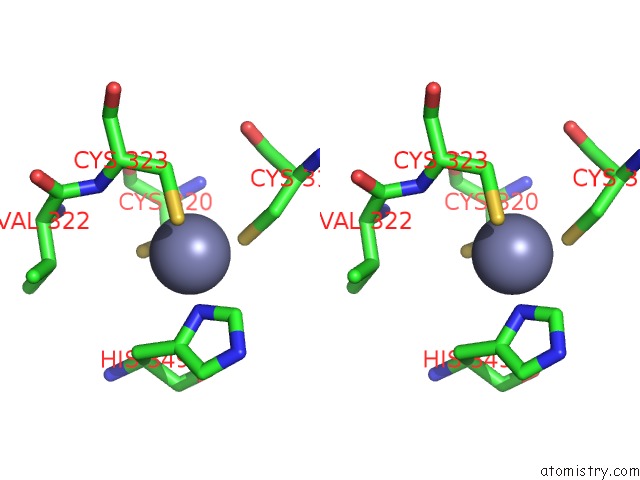
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Trna-Gunanine-Transglycosylase (Tgt) Mutant Y106F, C158V, A232S, V233G- Apo-Structure within 5.0Å range:
|
Reference:
I.Biela,
N.Tidten-Luksch,
F.Immekus,
S.Glinca,
T.X.Nguyen,
H.D.Gerber,
A.Heine,
G.Klebe,
K.Reuter.
Investigation of Specificity Determinants in Bacterial Trna-Guanine Transglycosylase Reveals Queuine, the Substrate of Its Eucaryotic Counterpart, As Inhibitor. Plos One V. 8 64240 2013.
ISSN: ESSN 1932-6203
PubMed: 23704982
DOI: 10.1371/JOURNAL.PONE.0064240
Page generated: Thu Oct 17 02:17:18 2024
ISSN: ESSN 1932-6203
PubMed: 23704982
DOI: 10.1371/JOURNAL.PONE.0064240
Last articles
K in 4ZUPK in 4ZXH
K in 4ZVJ
K in 4ZUN
K in 4ZUO
K in 4ZUQ
K in 4ZQR
K in 4ZUM
K in 4ZBM
K in 4ZN8