Zinc in PDB 9e8l: NUB1/FAT10-Processing Human 26S Proteasome Bound to TXNL1 with RPT4 at Top of Spiral Staircase
Enzymatic activity of NUB1/FAT10-Processing Human 26S Proteasome Bound to TXNL1 with RPT4 at Top of Spiral Staircase
All present enzymatic activity of NUB1/FAT10-Processing Human 26S Proteasome Bound to TXNL1 with RPT4 at Top of Spiral Staircase:
3.4.25.1;
3.4.25.1;
Other elements in 9e8l:
The structure of NUB1/FAT10-Processing Human 26S Proteasome Bound to TXNL1 with RPT4 at Top of Spiral Staircase also contains other interesting chemical elements:
| Magnesium | (Mg) | 3 atoms |
Zinc Binding Sites:
The binding sites of Zinc atom in the NUB1/FAT10-Processing Human 26S Proteasome Bound to TXNL1 with RPT4 at Top of Spiral Staircase
(pdb code 9e8l). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total only one binding site of Zinc was determined in the NUB1/FAT10-Processing Human 26S Proteasome Bound to TXNL1 with RPT4 at Top of Spiral Staircase, PDB code: 9e8l:
In total only one binding site of Zinc was determined in the NUB1/FAT10-Processing Human 26S Proteasome Bound to TXNL1 with RPT4 at Top of Spiral Staircase, PDB code: 9e8l:
Zinc binding site 1 out of 1 in 9e8l
Go back to
Zinc binding site 1 out
of 1 in the NUB1/FAT10-Processing Human 26S Proteasome Bound to TXNL1 with RPT4 at Top of Spiral Staircase
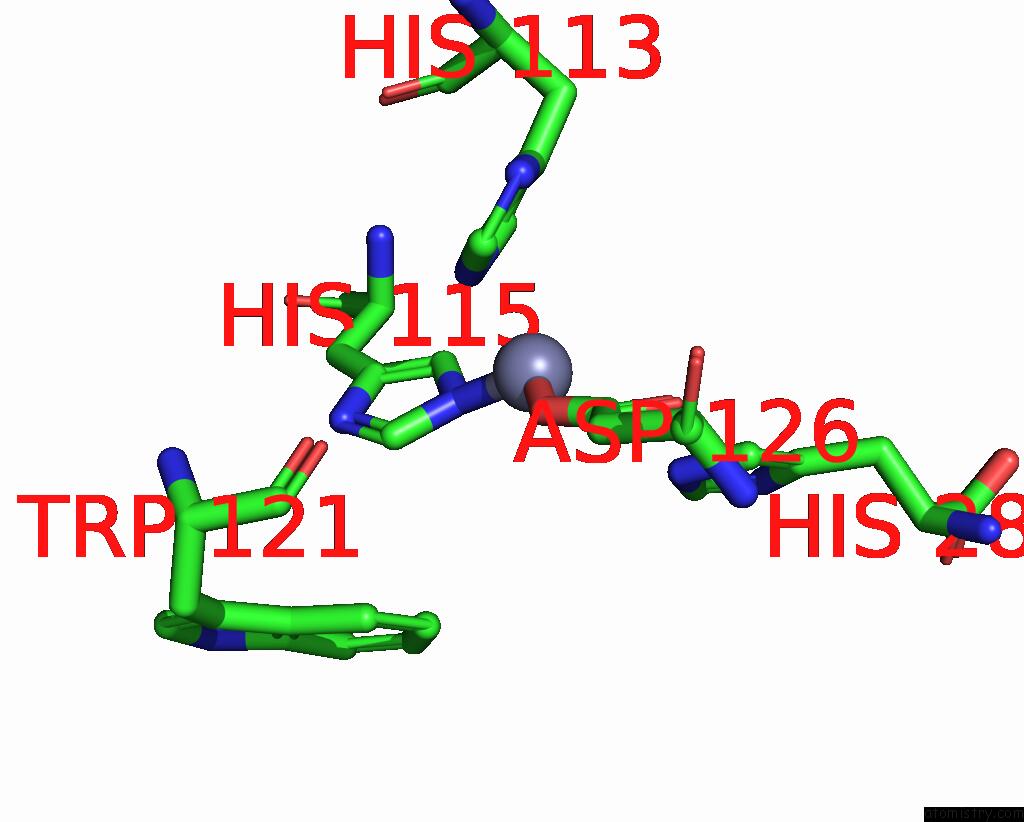
Mono view
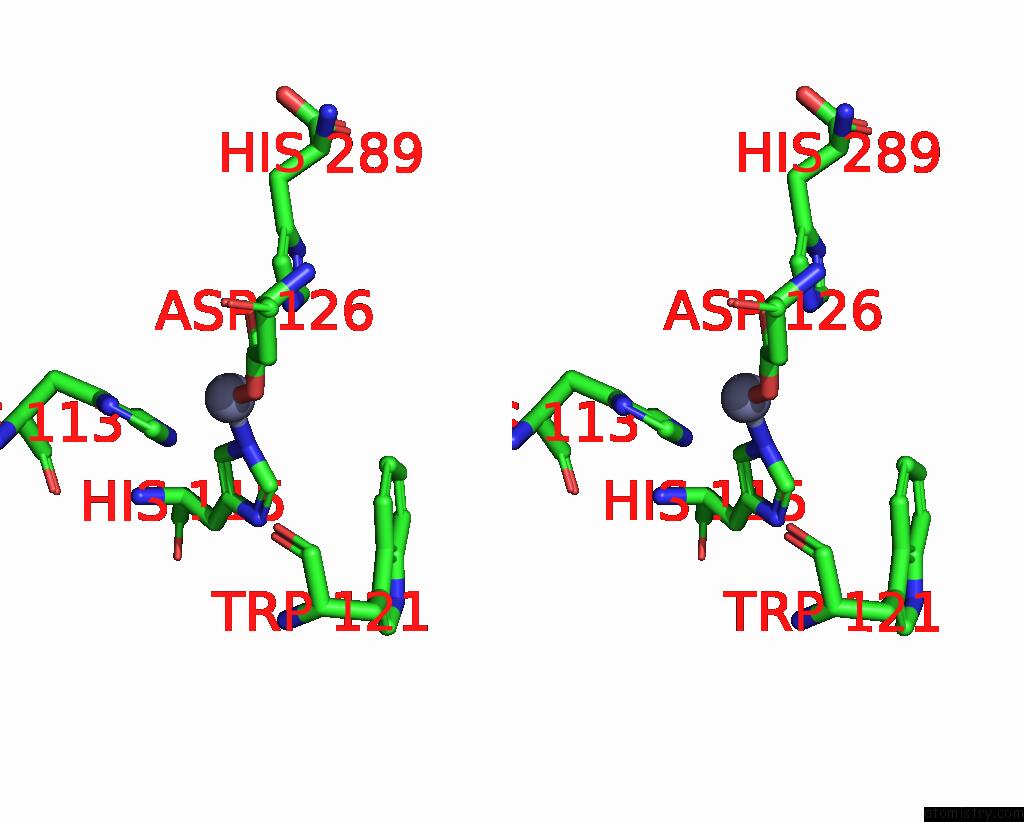
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of NUB1/FAT10-Processing Human 26S Proteasome Bound to TXNL1 with RPT4 at Top of Spiral Staircase within 5.0Å range:
|
Reference:
C.Arkinson,
C.L.Gee,
Z.Zhang,
K.C.Dong,
A.Martin.
Structural Landscape of Aaa+ Atpase Motor States in the Substrate-Degrading Human 26S Proteasome Reveals Conformation-Specific Binding of TXNL1. Biorxiv 2024.
ISSN: ISSN 2692-8205
PubMed: 39574680
DOI: 10.1101/2024.11.08.622731
Page generated: Tue Dec 10 21:56:08 2024
ISSN: ISSN 2692-8205
PubMed: 39574680
DOI: 10.1101/2024.11.08.622731
Last articles
Zn in 9MJ5Zn in 9HNW
Zn in 9G0L
Zn in 9FNE
Zn in 9DZN
Zn in 9E0I
Zn in 9D32
Zn in 9DAK
Zn in 8ZXC
Zn in 8ZUF