Zinc in PDB 9cre: Cryo-Em Structure of Sars-Cov-2 Spike Proteins on Intact Virions: Alpha (B.1.1.7) Variant 3 Closed Rbds
Zinc Binding Sites:
The binding sites of Zinc atom in the Cryo-Em Structure of Sars-Cov-2 Spike Proteins on Intact Virions: Alpha (B.1.1.7) Variant 3 Closed Rbds
(pdb code 9cre). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total only one binding site of Zinc was determined in the Cryo-Em Structure of Sars-Cov-2 Spike Proteins on Intact Virions: Alpha (B.1.1.7) Variant 3 Closed Rbds, PDB code: 9cre:
In total only one binding site of Zinc was determined in the Cryo-Em Structure of Sars-Cov-2 Spike Proteins on Intact Virions: Alpha (B.1.1.7) Variant 3 Closed Rbds, PDB code: 9cre:
Zinc binding site 1 out of 1 in 9cre
Go back to
Zinc binding site 1 out
of 1 in the Cryo-Em Structure of Sars-Cov-2 Spike Proteins on Intact Virions: Alpha (B.1.1.7) Variant 3 Closed Rbds
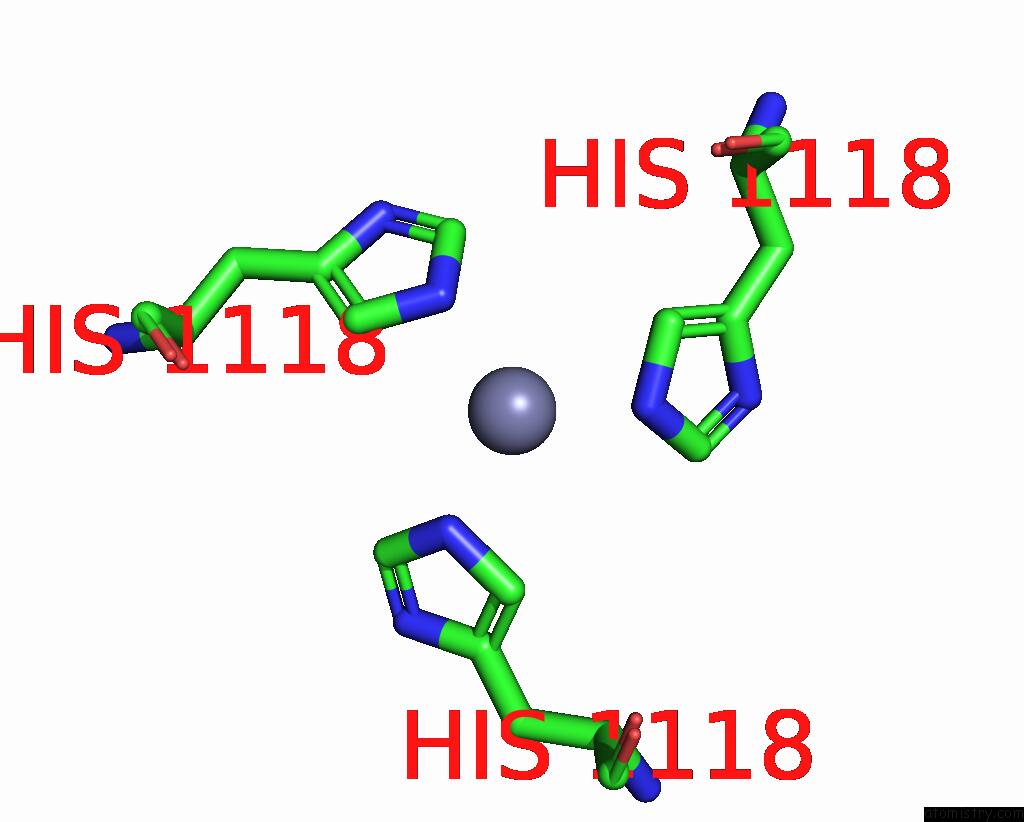
Mono view
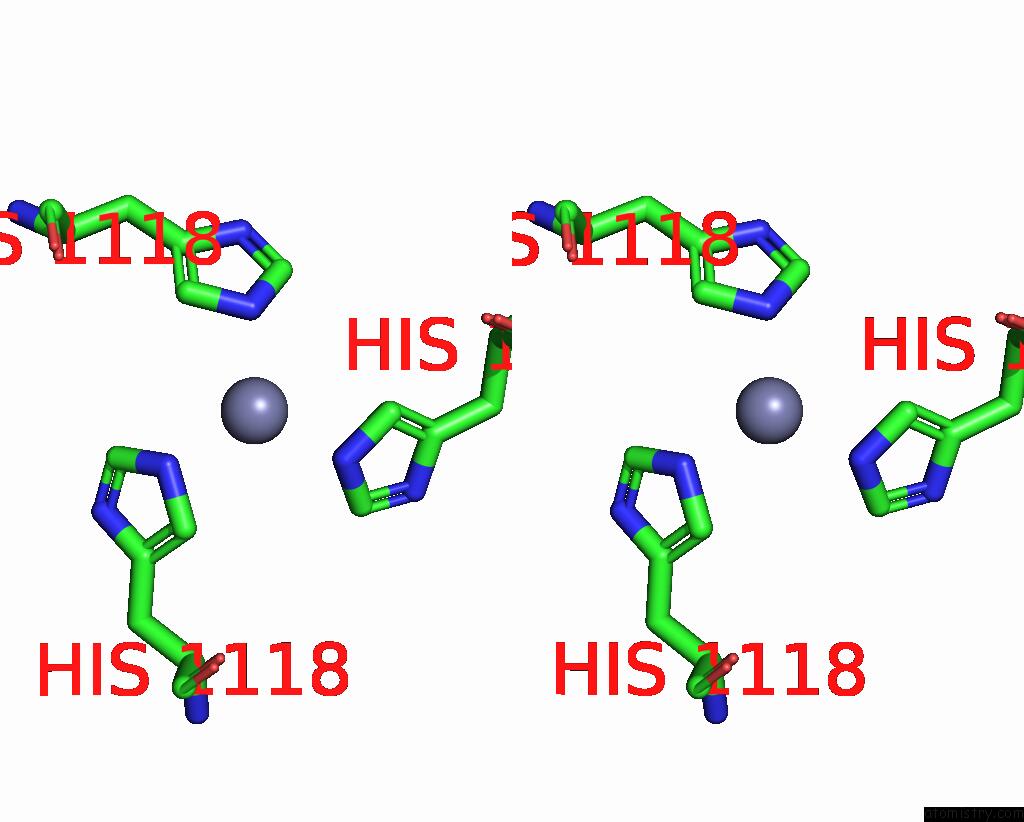
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Cryo-Em Structure of Sars-Cov-2 Spike Proteins on Intact Virions: Alpha (B.1.1.7) Variant 3 Closed Rbds within 5.0Å range:
|
Reference:
Z.Ke,
T.P.Peacock,
J.C.Brown,
C.M.Sheppard,
T.I.Croll,
A.Kotecha,
D.H.Goldhill,
W.S.Barclay,
J.A.G.Briggs.
Virion Morphology and on-Virus Spike Protein Structures of Diverse Sars-Cov-2 Variants. Embo J. 2024.
ISSN: ESSN 1460-2075
PubMed: 39543395
DOI: 10.1038/S44318-024-00303-1
Page generated: Tue Dec 10 21:56:08 2024
ISSN: ESSN 1460-2075
PubMed: 39543395
DOI: 10.1038/S44318-024-00303-1
Last articles
Zn in 9MJ5Zn in 9HNW
Zn in 9G0L
Zn in 9FNE
Zn in 9DZN
Zn in 9E0I
Zn in 9D32
Zn in 9DAK
Zn in 8ZXC
Zn in 8ZUF