Zinc »
PDB 8x3w-8xld »
8xbs »
Zinc in PDB 8xbs: C. Elegans Apo-SID1 Structure
Zinc Binding Sites:
The binding sites of Zinc atom in the C. Elegans Apo-SID1 Structure
(pdb code 8xbs). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 2 binding sites of Zinc where determined in the C. Elegans Apo-SID1 Structure, PDB code: 8xbs:
Jump to Zinc binding site number: 1; 2;
In total 2 binding sites of Zinc where determined in the C. Elegans Apo-SID1 Structure, PDB code: 8xbs:
Jump to Zinc binding site number: 1; 2;
Zinc binding site 1 out of 2 in 8xbs
Go back to
Zinc binding site 1 out
of 2 in the C. Elegans Apo-SID1 Structure
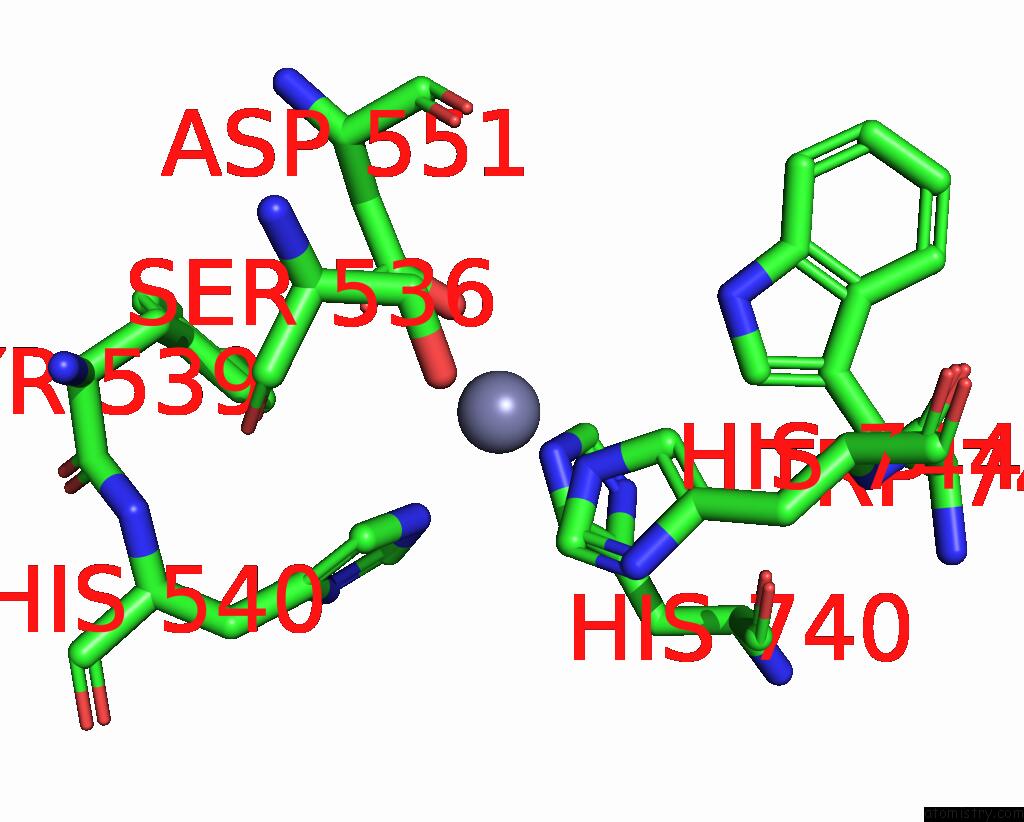
Mono view
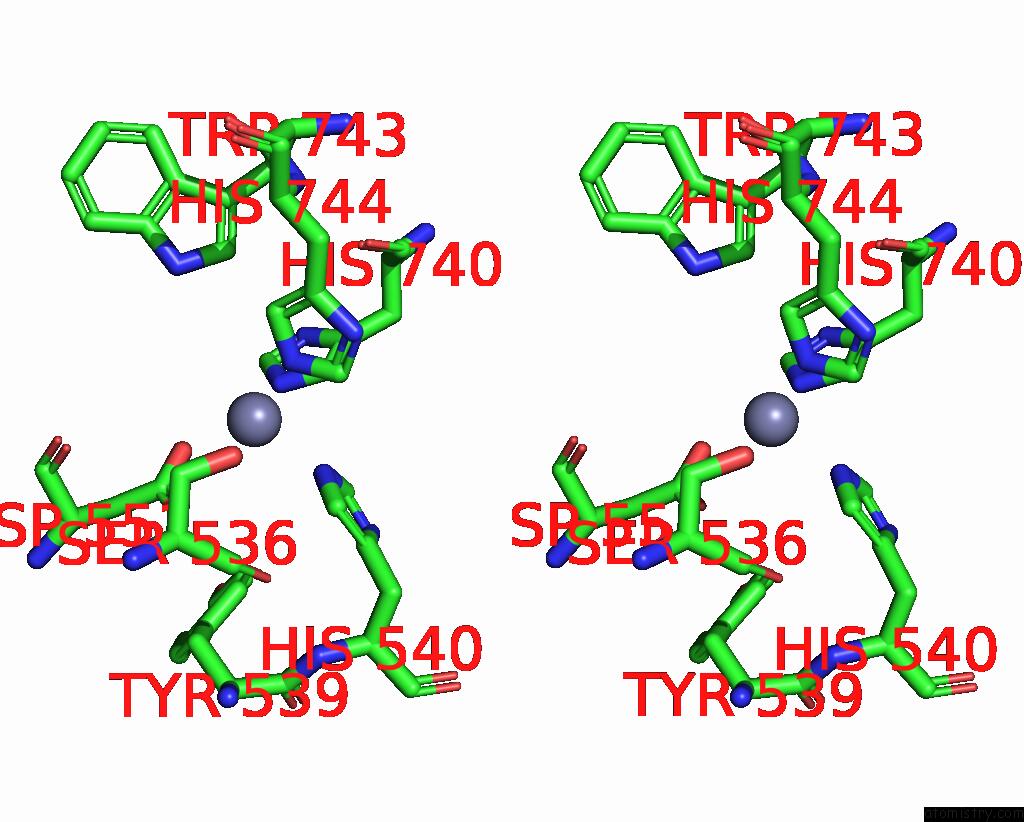
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of C. Elegans Apo-SID1 Structure within 5.0Å range:
|
Zinc binding site 2 out of 2 in 8xbs
Go back to
Zinc binding site 2 out
of 2 in the C. Elegans Apo-SID1 Structure
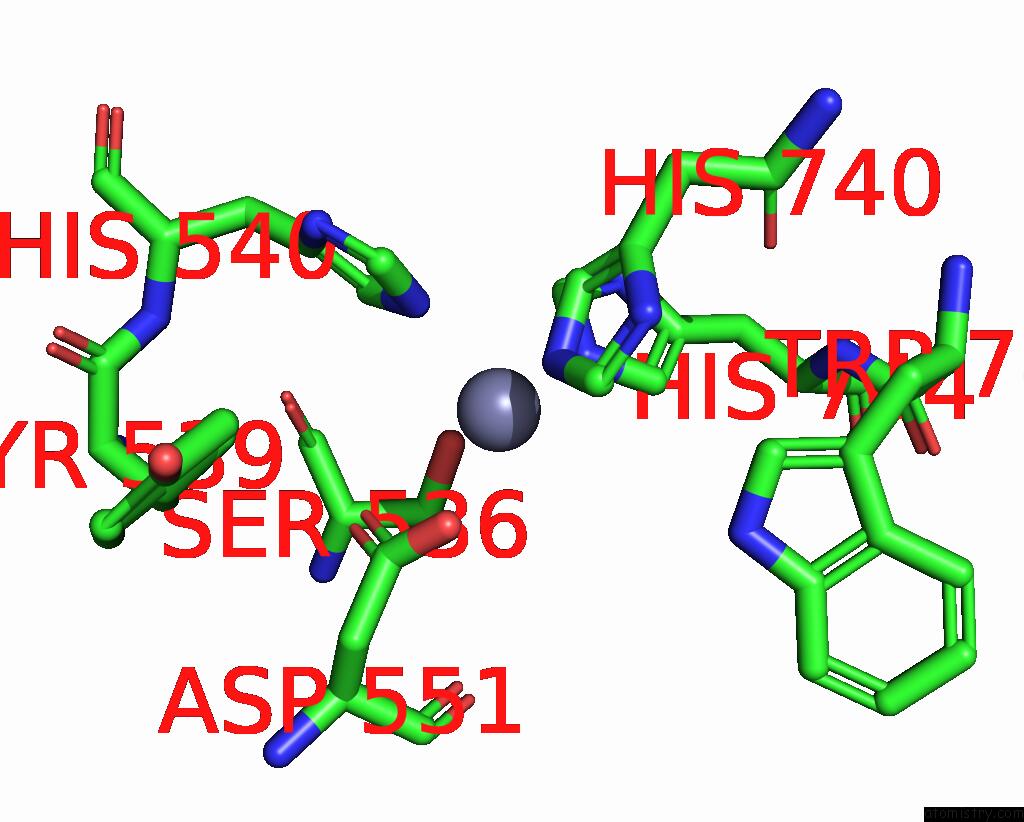
Mono view
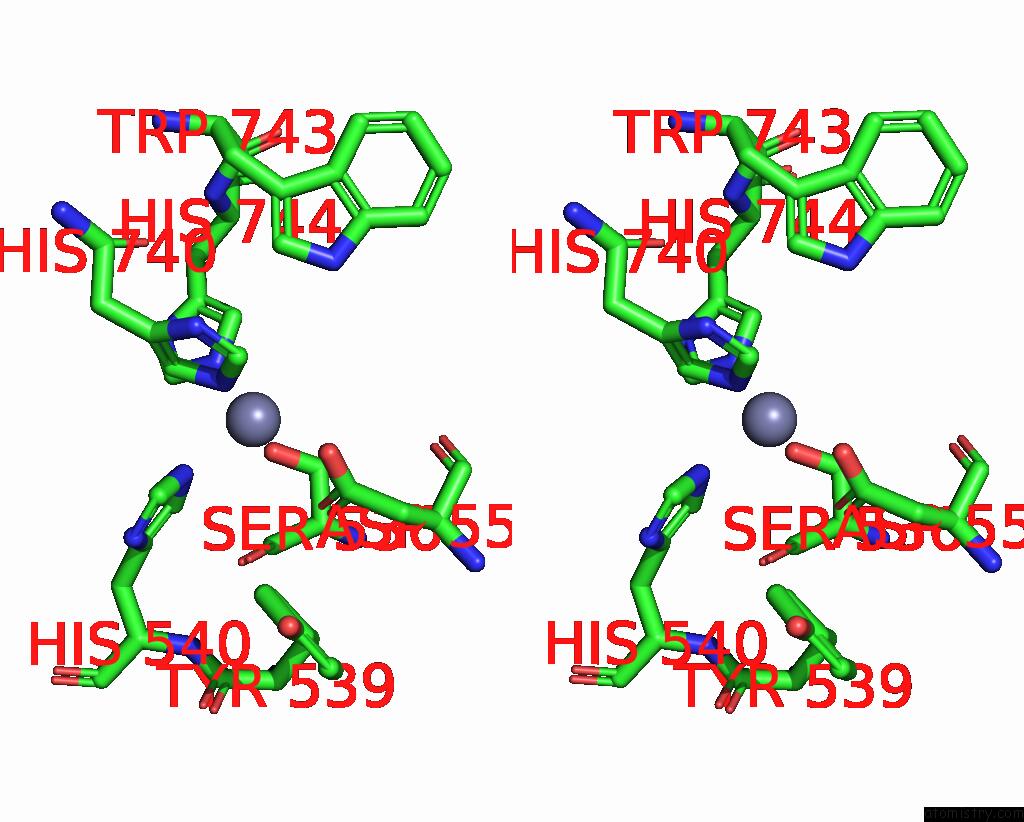
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of C. Elegans Apo-SID1 Structure within 5.0Å range:
|
Reference:
R.Wang,
Y.Cong,
D.Qian,
C.Yan,
D.Gong.
Structural Basis For Double-Stranded Rna Recognition By SID1. Nucleic Acids Res. 2024.
ISSN: ESSN 1362-4962
PubMed: 38742627
DOI: 10.1093/NAR/GKAE395
Page generated: Fri Aug 22 15:27:21 2025
ISSN: ESSN 1362-4962
PubMed: 38742627
DOI: 10.1093/NAR/GKAE395
Last articles
Zr in 1XC1Zr in 6Y7P
Zr in 6GNL
Zr in 6HYB
Zr in 4XYY
Zr in 5KHP
Zn in 9VXG
Zn in 9VWY
Zn in 9VCL
Zn in 9VKN