Zinc »
PDB 8we0-8wo3 »
8wlr »
Zinc in PDB 8wlr: Cryo-Em Structure of Sars-Cov-2 Prototype Spike Protein Receptor- Binding Domain in Complex with Hippopotamus ACE2
Zinc Binding Sites:
The binding sites of Zinc atom in the Cryo-Em Structure of Sars-Cov-2 Prototype Spike Protein Receptor- Binding Domain in Complex with Hippopotamus ACE2
(pdb code 8wlr). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total only one binding site of Zinc was determined in the Cryo-Em Structure of Sars-Cov-2 Prototype Spike Protein Receptor- Binding Domain in Complex with Hippopotamus ACE2, PDB code: 8wlr:
In total only one binding site of Zinc was determined in the Cryo-Em Structure of Sars-Cov-2 Prototype Spike Protein Receptor- Binding Domain in Complex with Hippopotamus ACE2, PDB code: 8wlr:
Zinc binding site 1 out of 1 in 8wlr
Go back to
Zinc binding site 1 out
of 1 in the Cryo-Em Structure of Sars-Cov-2 Prototype Spike Protein Receptor- Binding Domain in Complex with Hippopotamus ACE2
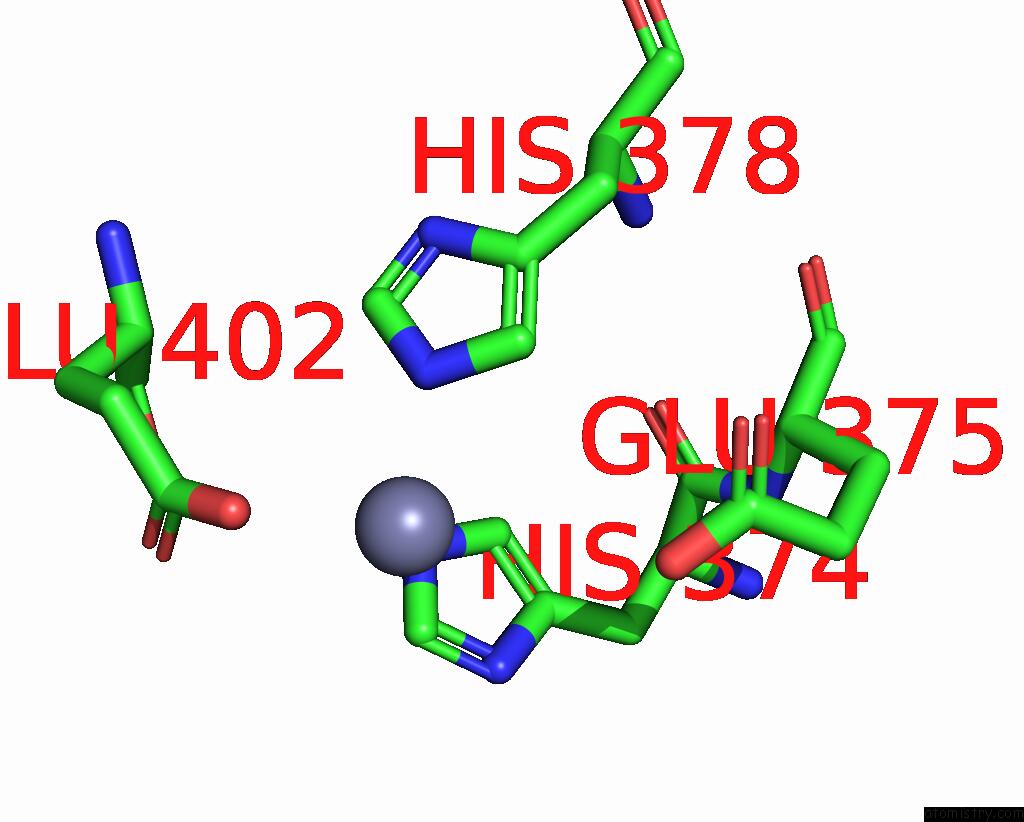
Mono view
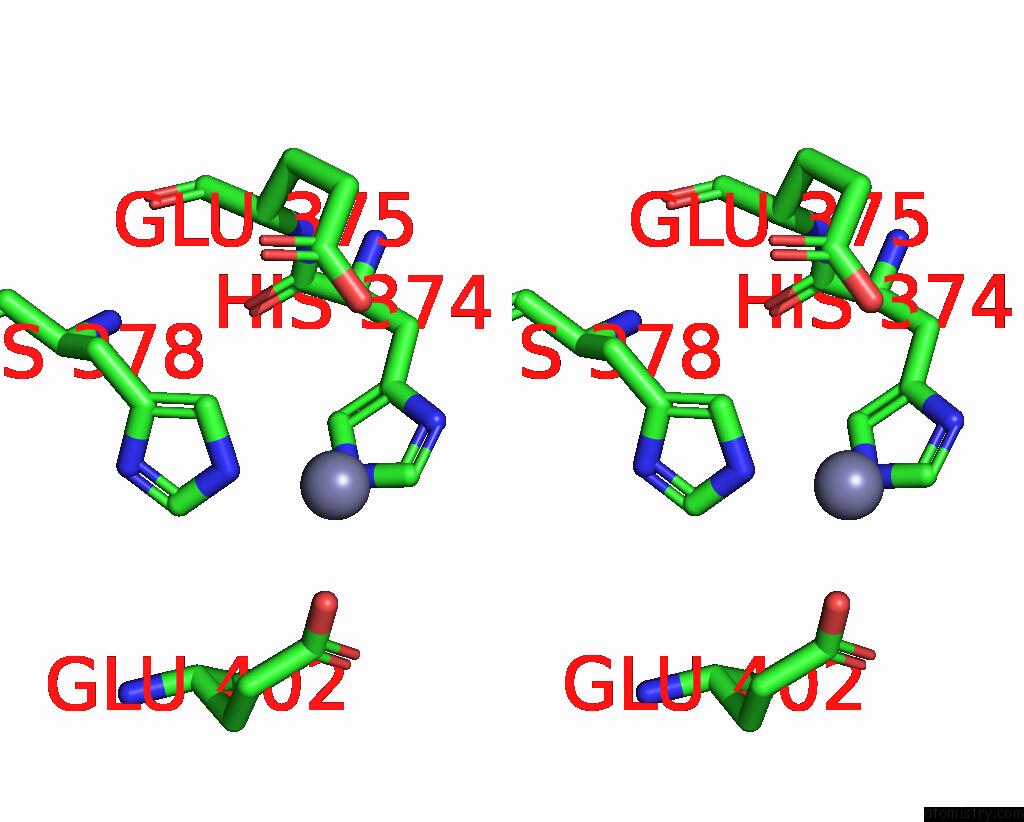
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Cryo-Em Structure of Sars-Cov-2 Prototype Spike Protein Receptor- Binding Domain in Complex with Hippopotamus ACE2 within 5.0Å range:
|
Reference:
R.Yang,
P.Han,
P.Han,
D.Li,
R.Zhao,
S.Niu,
K.Liu,
S.Li,
W.Tian,
G.F.Gao.
Molecular Basis of Hippopotamus ACE2 Binding to Sars-Cov-2 To Be Published.
Page generated: Fri Aug 22 15:13:21 2025
Last articles
Zr in 1XC1Zr in 6Y7P
Zr in 6GNL
Zr in 6HYB
Zr in 4XYY
Zr in 5KHP
Zn in 9VXG
Zn in 9VWY
Zn in 9VCL
Zn in 9VKN