Zinc »
PDB 8w40-8wdz »
8w6r »
Zinc in PDB 8w6r: Murine SMPDL3A Bound to Sulfate
Protein crystallography data
The structure of Murine SMPDL3A Bound to Sulfate, PDB code: 8w6r
was solved by
C.Zhang,
P.Liu,
S.Fan,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 20.54 / 1.95 |
| Space group | P 41 |
| Cell size a, b, c (Å), α, β, γ (°) | 86.374, 86.374, 79.886, 90, 90, 90 |
| R / Rfree (%) | 22.1 / 25.2 |
Zinc Binding Sites:
The binding sites of Zinc atom in the Murine SMPDL3A Bound to Sulfate
(pdb code 8w6r). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 2 binding sites of Zinc where determined in the Murine SMPDL3A Bound to Sulfate, PDB code: 8w6r:
Jump to Zinc binding site number: 1; 2;
In total 2 binding sites of Zinc where determined in the Murine SMPDL3A Bound to Sulfate, PDB code: 8w6r:
Jump to Zinc binding site number: 1; 2;
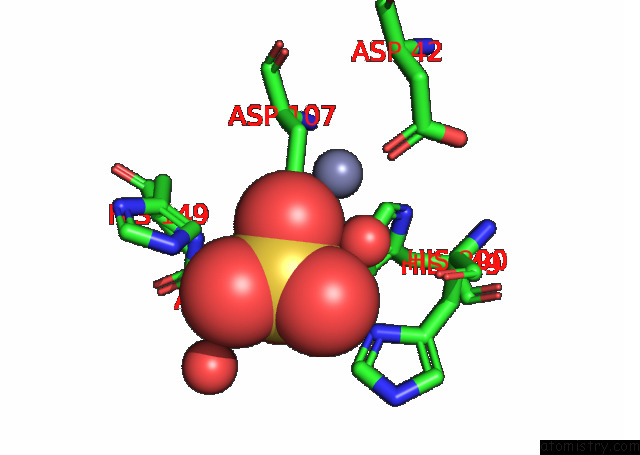
Zinc binding site 1 out of 2 in 8w6r
Go back to
Zinc binding site 1 out
of 2 in the Murine SMPDL3A Bound to Sulfate
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Murine SMPDL3A Bound to Sulfate within 5.0Å range:
|
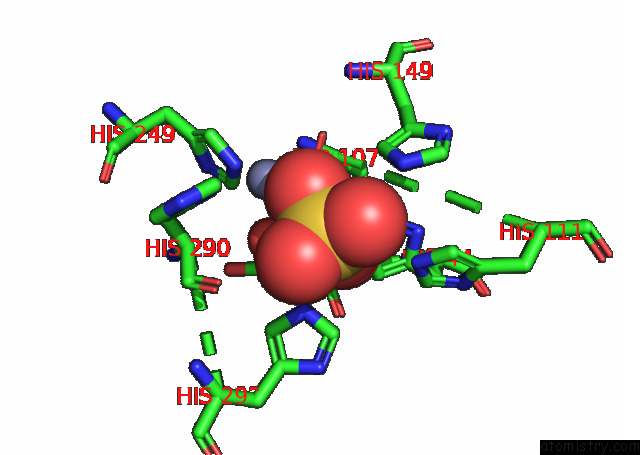
Zinc binding site 2 out of 2 in 8w6r
Go back to
Zinc binding site 2 out
of 2 in the Murine SMPDL3A Bound to Sulfate
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of Murine SMPDL3A Bound to Sulfate within 5.0Å range:
|
Reference:
Y.Hou,
Z.Wang,
P.Liu,
X.Wei,
Z.Zhang,
S.Fan,
L.Zhang,
F.Han,
Y.Song,
L.Chu,
C.Zhang.
SMPDL3A Is A Cgamp-Degrading Enzyme Induced By Lxr-Mediated Lipid Metabolism to Restrict Cgas-Sting Dna Sensing. Immunity V. 56 2492 2023.
ISSN: ISSN 1074-7613
PubMed: 37890481
DOI: 10.1016/J.IMMUNI.2023.10.001
Page generated: Fri Aug 22 15:00:18 2025
ISSN: ISSN 1074-7613
PubMed: 37890481
DOI: 10.1016/J.IMMUNI.2023.10.001
Last articles
Zr in 1XC1Zr in 6Y7P
Zr in 6GNL
Zr in 6HYB
Zr in 4XYY
Zr in 5KHP
Zn in 9VXG
Zn in 9VWY
Zn in 9VCL
Zn in 9VKN