Zinc in PDB 8vaq: Structure of the E. Coli Clamp Loader Bound to the Beta Clamp in A Closed-DNA1 Conformation
Enzymatic activity of Structure of the E. Coli Clamp Loader Bound to the Beta Clamp in A Closed-DNA1 Conformation
All present enzymatic activity of Structure of the E. Coli Clamp Loader Bound to the Beta Clamp in A Closed-DNA1 Conformation:
2.7.7.7;
2.7.7.7;
Other elements in 8vaq:
The structure of Structure of the E. Coli Clamp Loader Bound to the Beta Clamp in A Closed-DNA1 Conformation also contains other interesting chemical elements:
| Magnesium | (Mg) | 3 atoms |
| Fluorine | (F) | 9 atoms |
Zinc Binding Sites:
The binding sites of Zinc atom in the Structure of the E. Coli Clamp Loader Bound to the Beta Clamp in A Closed-DNA1 Conformation
(pdb code 8vaq). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 4 binding sites of Zinc where determined in the Structure of the E. Coli Clamp Loader Bound to the Beta Clamp in A Closed-DNA1 Conformation, PDB code: 8vaq:
Jump to Zinc binding site number: 1; 2; 3; 4;
In total 4 binding sites of Zinc where determined in the Structure of the E. Coli Clamp Loader Bound to the Beta Clamp in A Closed-DNA1 Conformation, PDB code: 8vaq:
Jump to Zinc binding site number: 1; 2; 3; 4;
Zinc binding site 1 out of 4 in 8vaq
Go back to
Zinc binding site 1 out
of 4 in the Structure of the E. Coli Clamp Loader Bound to the Beta Clamp in A Closed-DNA1 Conformation
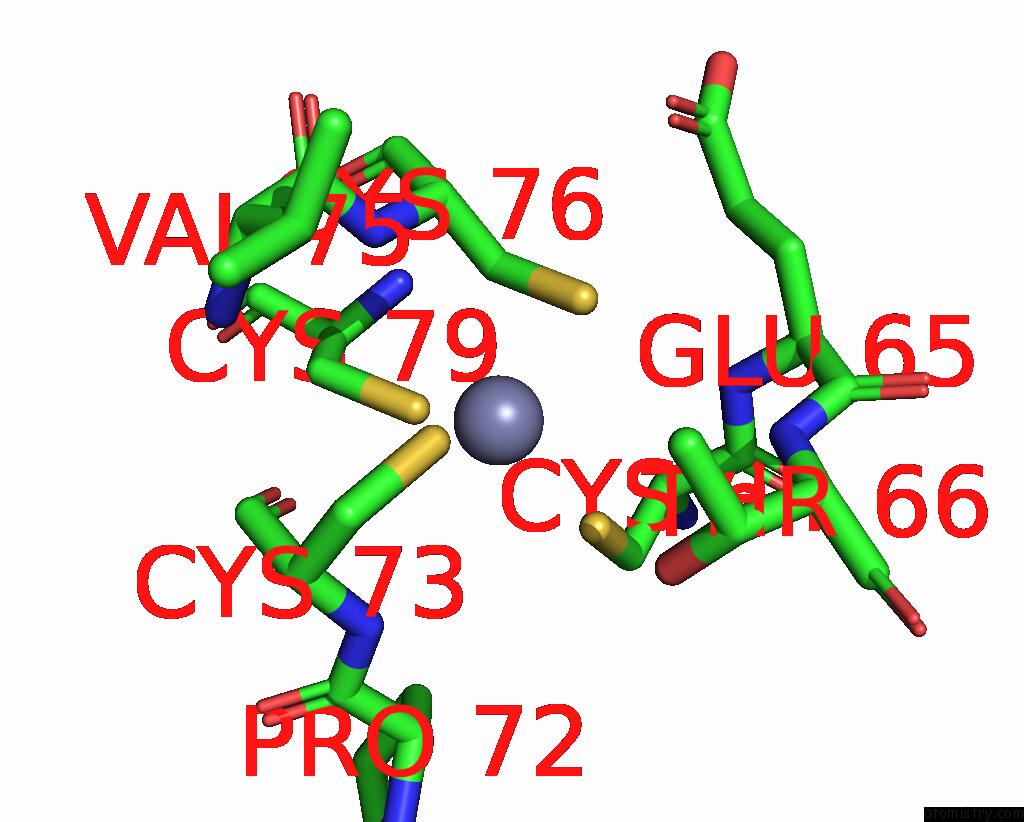
Mono view
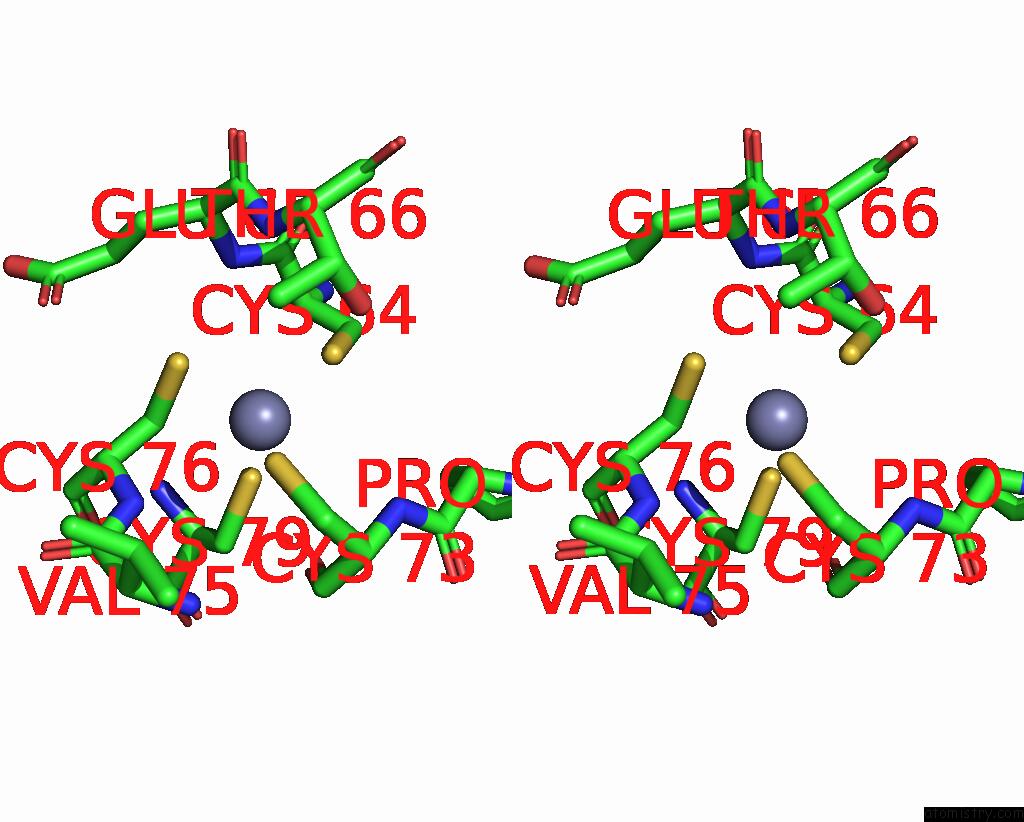
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Structure of the E. Coli Clamp Loader Bound to the Beta Clamp in A Closed-DNA1 Conformation within 5.0Å range:
|
Zinc binding site 2 out of 4 in 8vaq
Go back to
Zinc binding site 2 out
of 4 in the Structure of the E. Coli Clamp Loader Bound to the Beta Clamp in A Closed-DNA1 Conformation
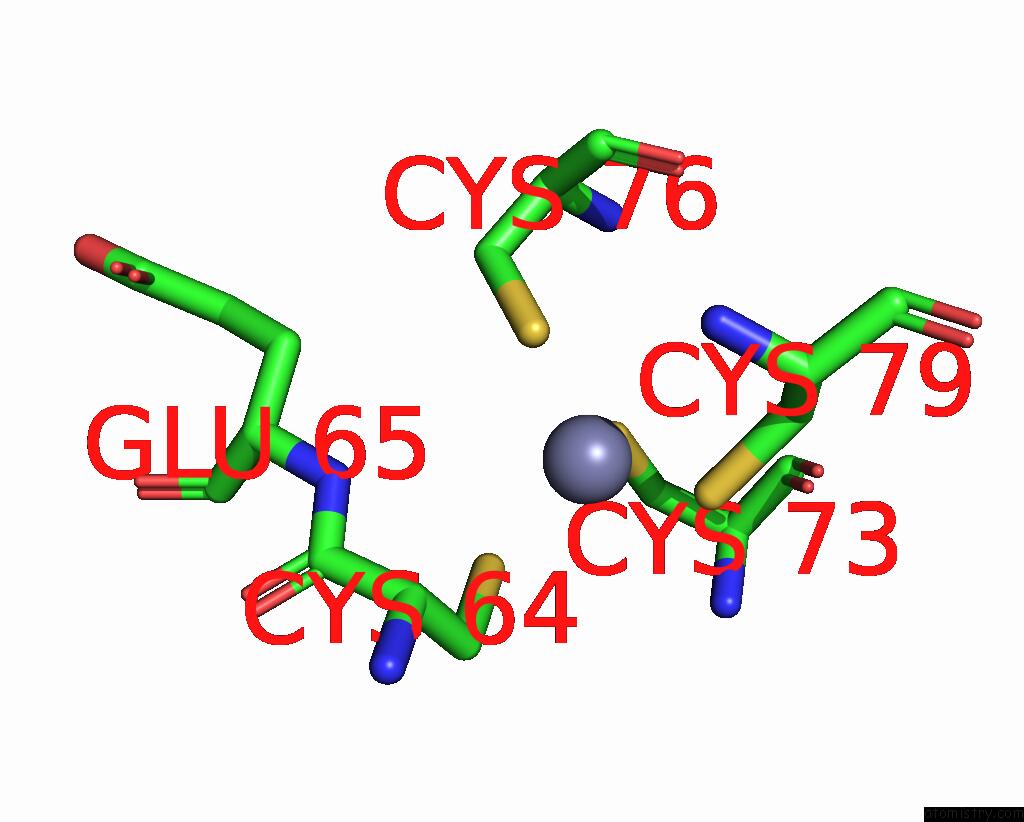
Mono view
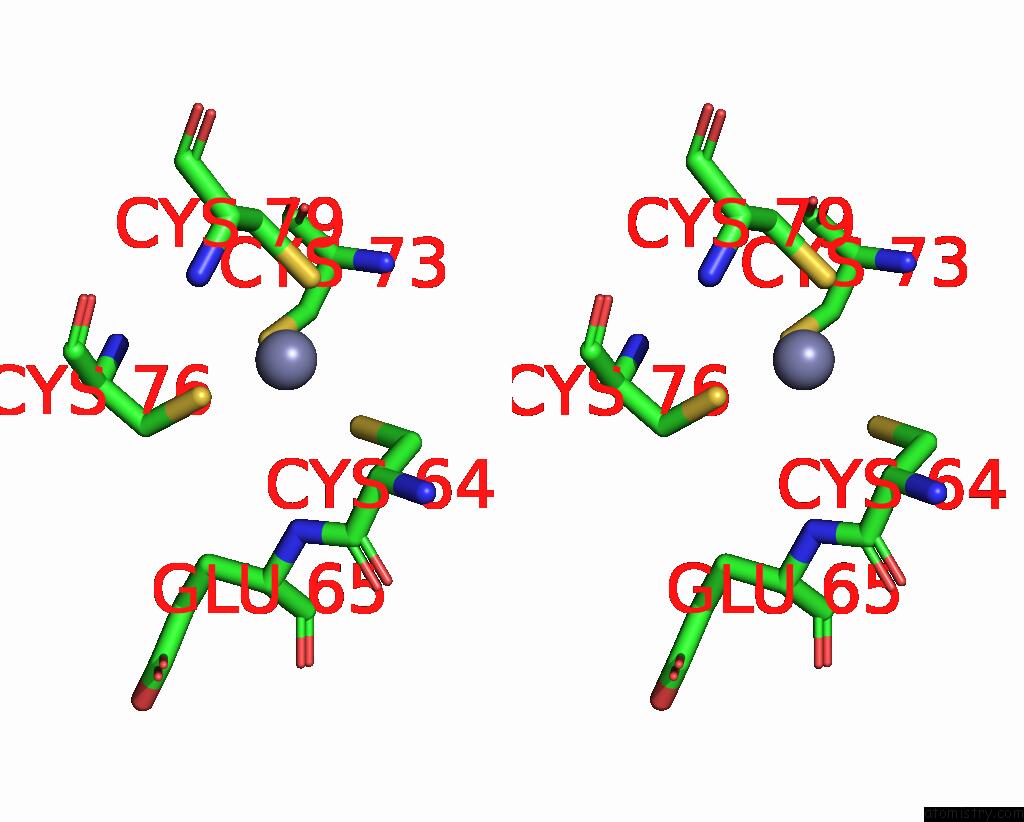
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of Structure of the E. Coli Clamp Loader Bound to the Beta Clamp in A Closed-DNA1 Conformation within 5.0Å range:
|
Zinc binding site 3 out of 4 in 8vaq
Go back to
Zinc binding site 3 out
of 4 in the Structure of the E. Coli Clamp Loader Bound to the Beta Clamp in A Closed-DNA1 Conformation
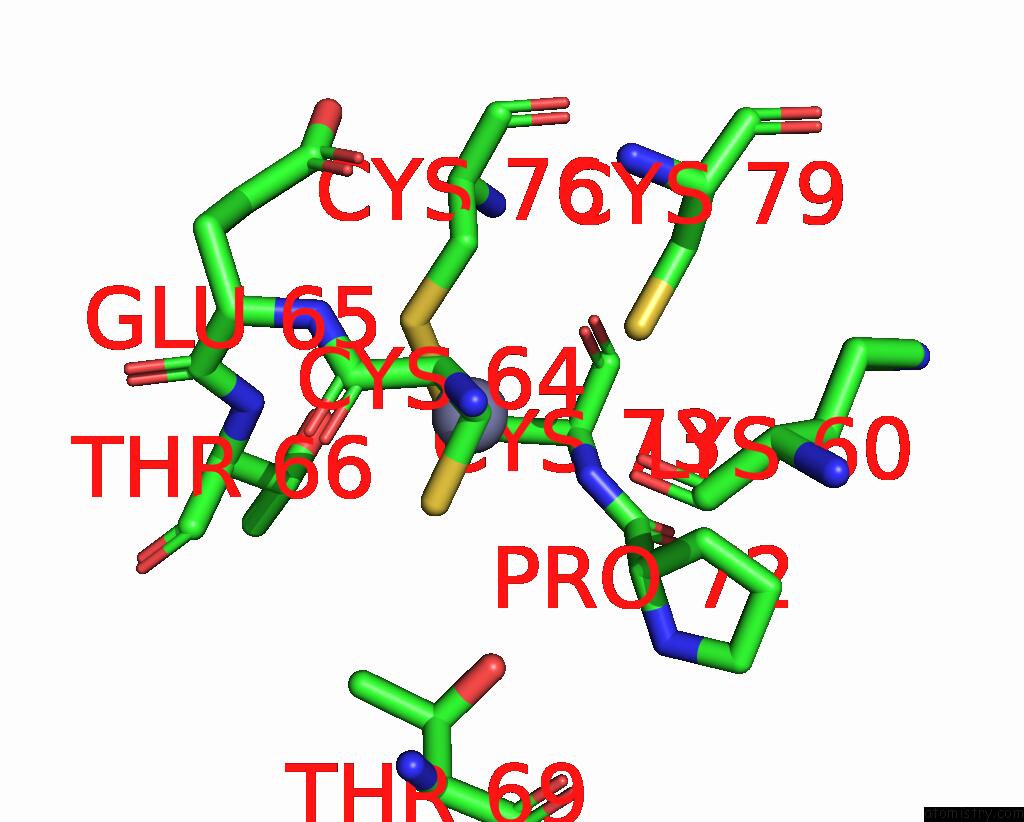
Mono view
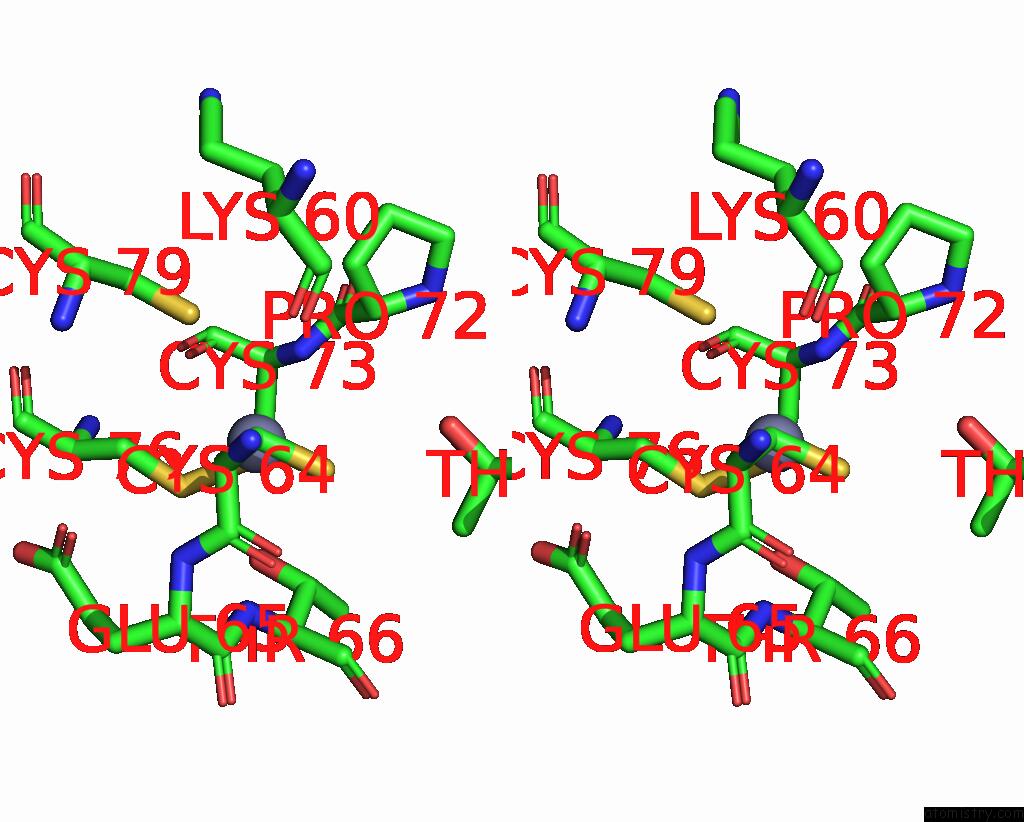
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 3 of Structure of the E. Coli Clamp Loader Bound to the Beta Clamp in A Closed-DNA1 Conformation within 5.0Å range:
|
Zinc binding site 4 out of 4 in 8vaq
Go back to
Zinc binding site 4 out
of 4 in the Structure of the E. Coli Clamp Loader Bound to the Beta Clamp in A Closed-DNA1 Conformation
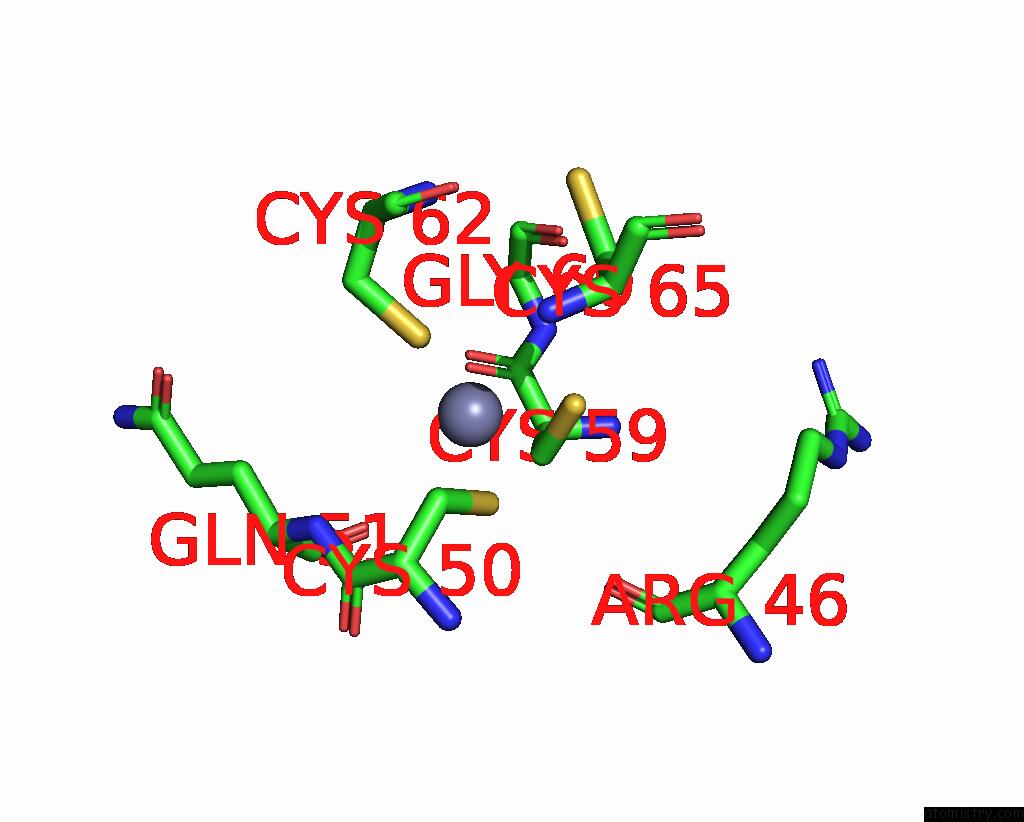
Mono view
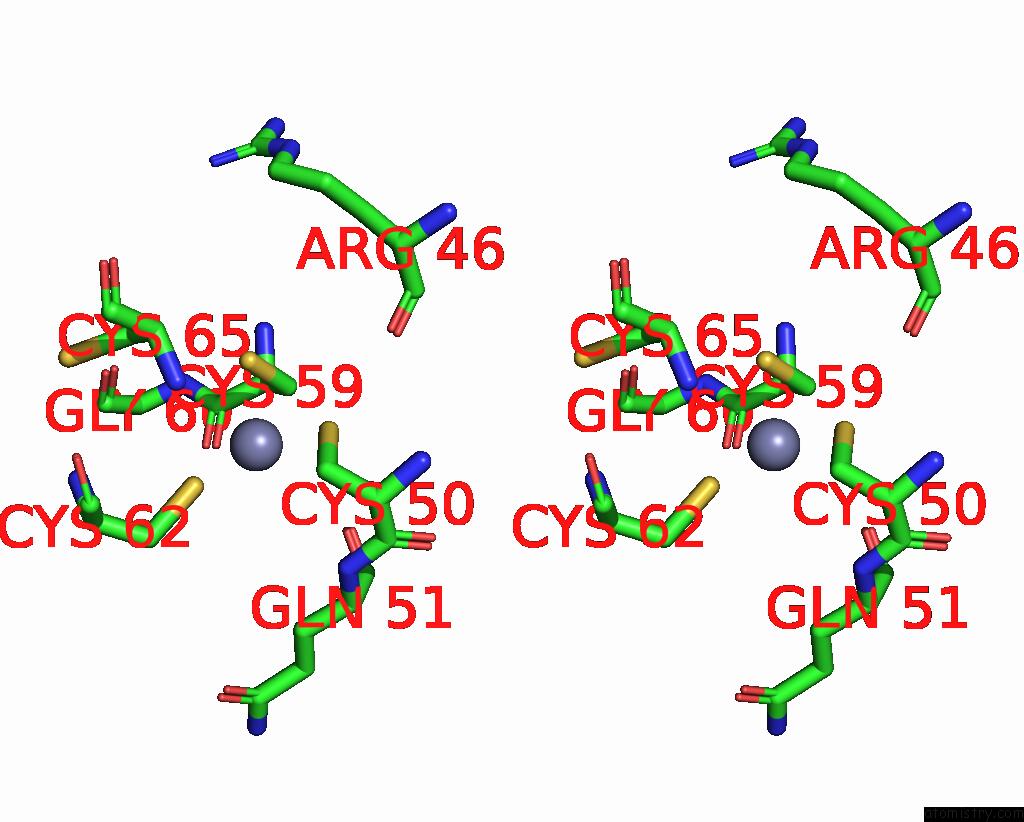
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 4 of Structure of the E. Coli Clamp Loader Bound to the Beta Clamp in A Closed-DNA1 Conformation within 5.0Å range:
|
Reference:
J.T.Landeck,
J.Pajak,
E.K.Norman,
E.L.Sedivy,
B.A.Kelch.
Differences Between Bacteria and Eukaryotes in Clamp Loader Mechanism, A Conserved Process Underlying Dna Replication. J.Biol.Chem. 07166 2024.
ISSN: ESSN 1083-351X
PubMed: 38490435
DOI: 10.1016/J.JBC.2024.107166
Page generated: Thu Oct 31 12:43:44 2024
ISSN: ESSN 1083-351X
PubMed: 38490435
DOI: 10.1016/J.JBC.2024.107166
Last articles
Zn in 9MJ5Zn in 9HNW
Zn in 9G0L
Zn in 9FNE
Zn in 9DZN
Zn in 9E0I
Zn in 9D32
Zn in 9DAK
Zn in 8ZXC
Zn in 8ZUF