Zinc »
PDB 8q1y-8qf5 »
8q7h »
Zinc in PDB 8q7h: Structure of CUL9-RBX1 Ubiquitin E3 Ligase Complex in Unneddylated and Neddylated Conformation - Focused Cullin Dimer
Enzymatic activity of Structure of CUL9-RBX1 Ubiquitin E3 Ligase Complex in Unneddylated and Neddylated Conformation - Focused Cullin Dimer
All present enzymatic activity of Structure of CUL9-RBX1 Ubiquitin E3 Ligase Complex in Unneddylated and Neddylated Conformation - Focused Cullin Dimer:
2.3.2.27; 2.3.2.32;
2.3.2.27; 2.3.2.32;
Zinc Binding Sites:
The binding sites of Zinc atom in the Structure of CUL9-RBX1 Ubiquitin E3 Ligase Complex in Unneddylated and Neddylated Conformation - Focused Cullin Dimer
(pdb code 8q7h). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 10 binding sites of Zinc where determined in the Structure of CUL9-RBX1 Ubiquitin E3 Ligase Complex in Unneddylated and Neddylated Conformation - Focused Cullin Dimer, PDB code: 8q7h:
Jump to Zinc binding site number: 1; 2; 3; 4; 5; 6; 7; 8; 9; 10;
In total 10 binding sites of Zinc where determined in the Structure of CUL9-RBX1 Ubiquitin E3 Ligase Complex in Unneddylated and Neddylated Conformation - Focused Cullin Dimer, PDB code: 8q7h:
Jump to Zinc binding site number: 1; 2; 3; 4; 5; 6; 7; 8; 9; 10;
Zinc binding site 1 out of 10 in 8q7h
Go back to
Zinc binding site 1 out
of 10 in the Structure of CUL9-RBX1 Ubiquitin E3 Ligase Complex in Unneddylated and Neddylated Conformation - Focused Cullin Dimer
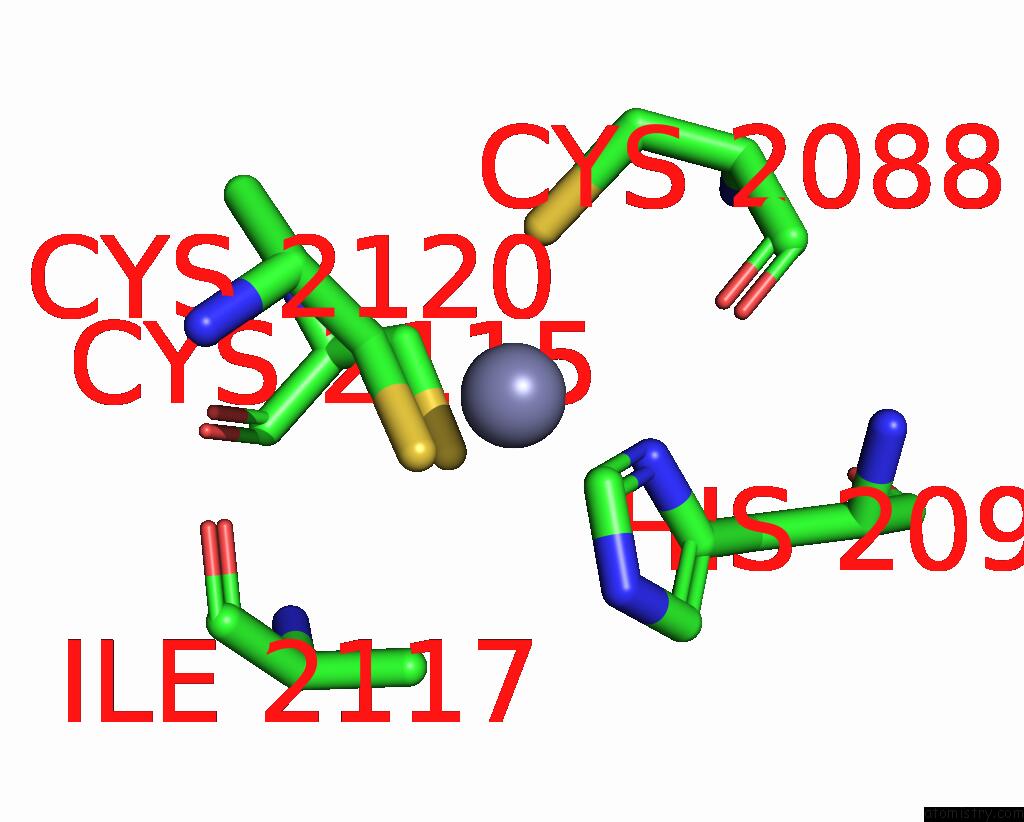
Mono view
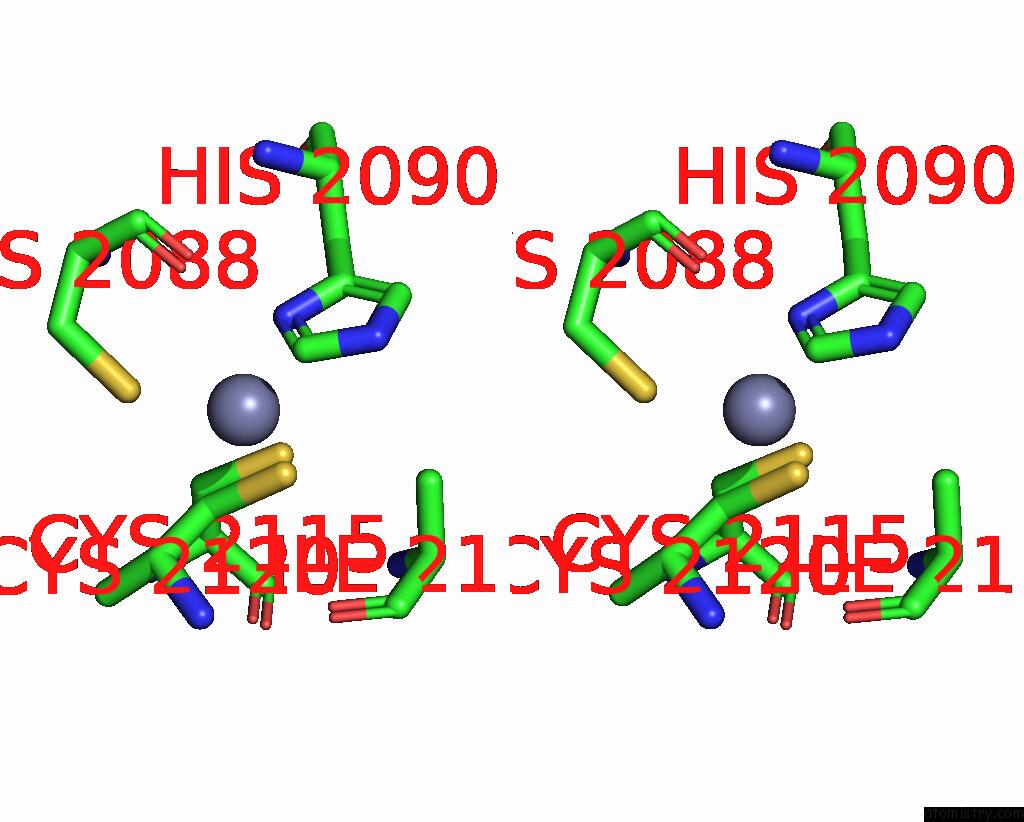
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Structure of CUL9-RBX1 Ubiquitin E3 Ligase Complex in Unneddylated and Neddylated Conformation - Focused Cullin Dimer within 5.0Å range:
|
Zinc binding site 2 out of 10 in 8q7h
Go back to
Zinc binding site 2 out
of 10 in the Structure of CUL9-RBX1 Ubiquitin E3 Ligase Complex in Unneddylated and Neddylated Conformation - Focused Cullin Dimer
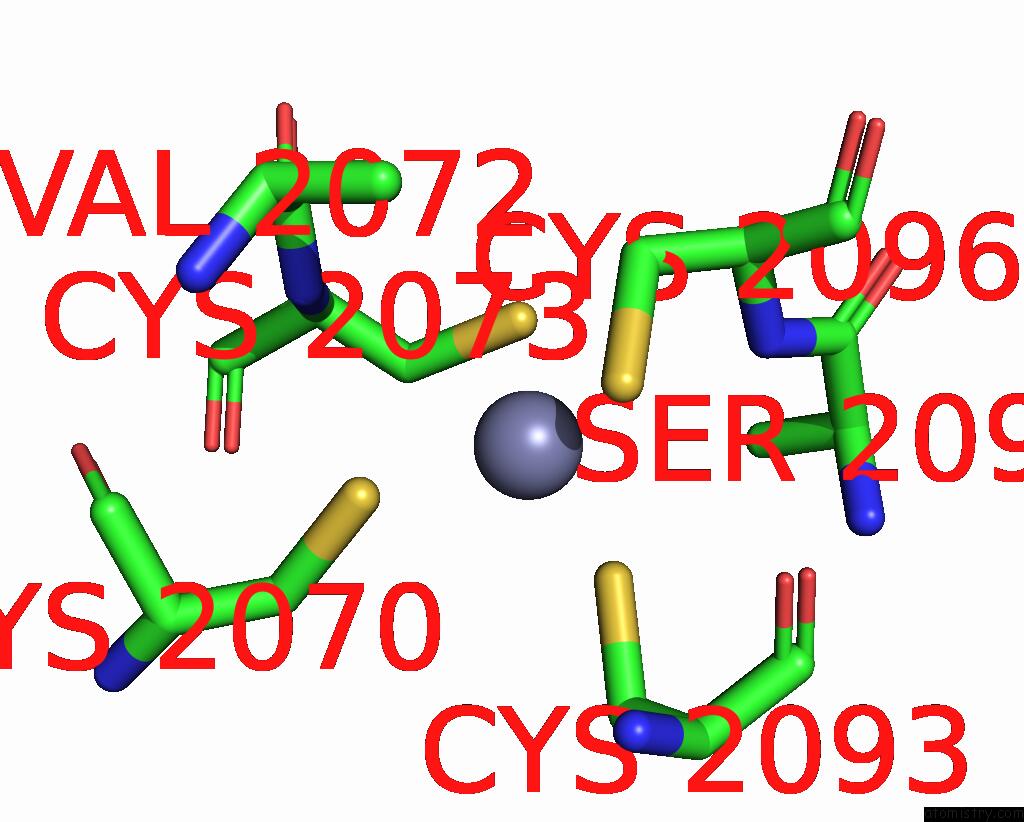
Mono view
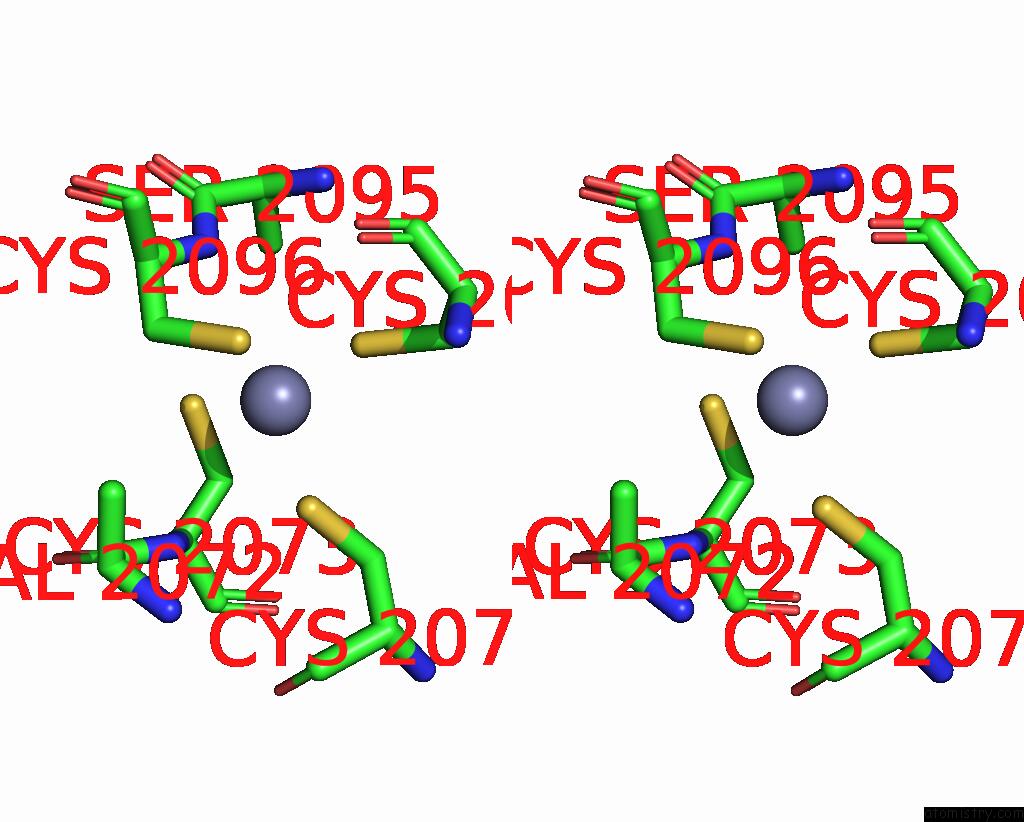
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of Structure of CUL9-RBX1 Ubiquitin E3 Ligase Complex in Unneddylated and Neddylated Conformation - Focused Cullin Dimer within 5.0Å range:
|
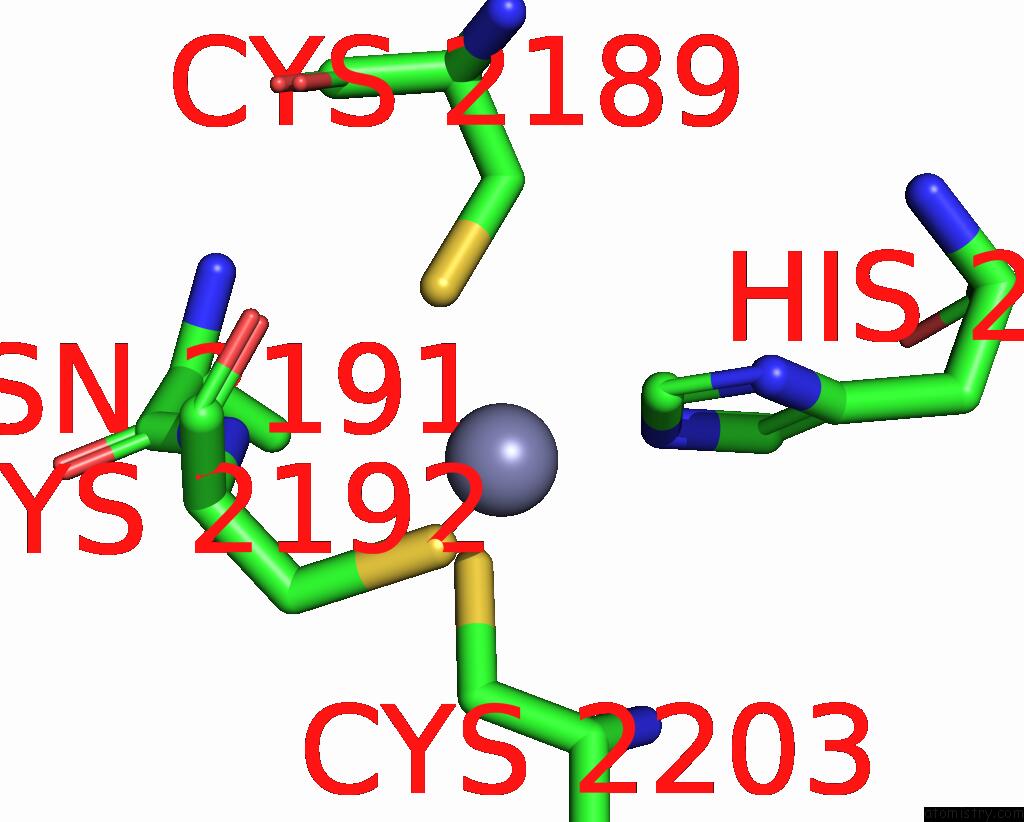
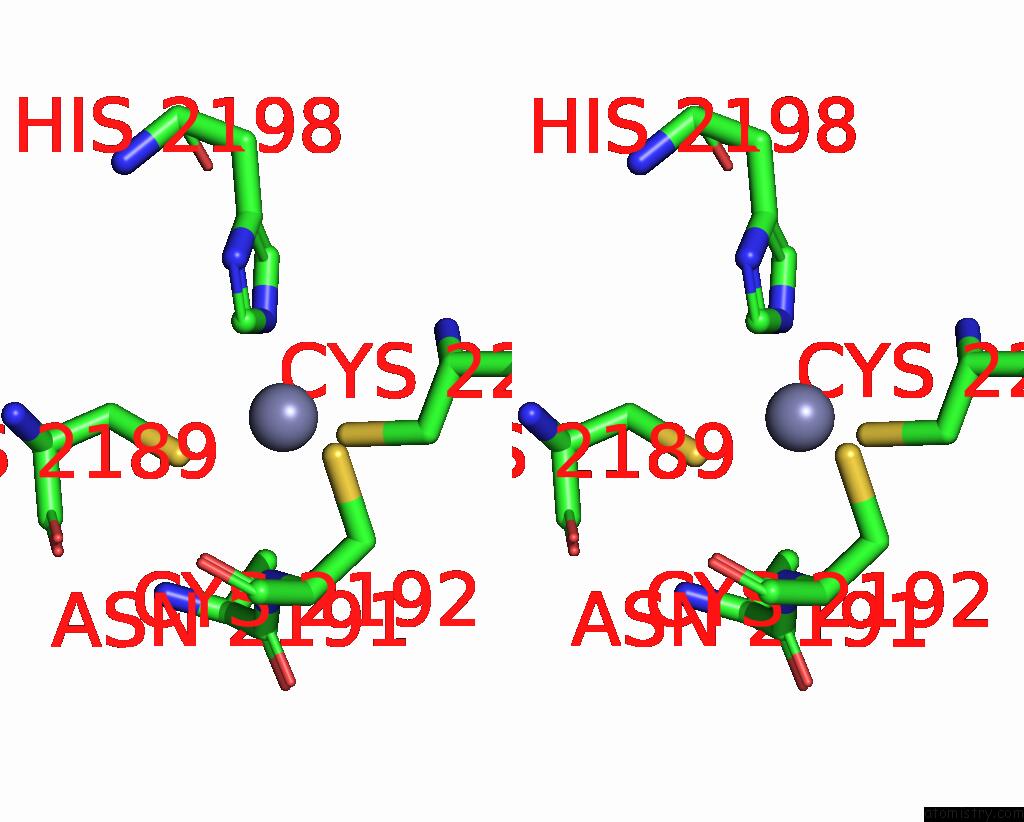
Zinc binding site 3 out of 10 in 8q7h
Go back to
Zinc binding site 3 out
of 10 in the Structure of CUL9-RBX1 Ubiquitin E3 Ligase Complex in Unneddylated and Neddylated Conformation - Focused Cullin Dimer
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 3 of Structure of CUL9-RBX1 Ubiquitin E3 Ligase Complex in Unneddylated and Neddylated Conformation - Focused Cullin Dimer within 5.0Å range:
|
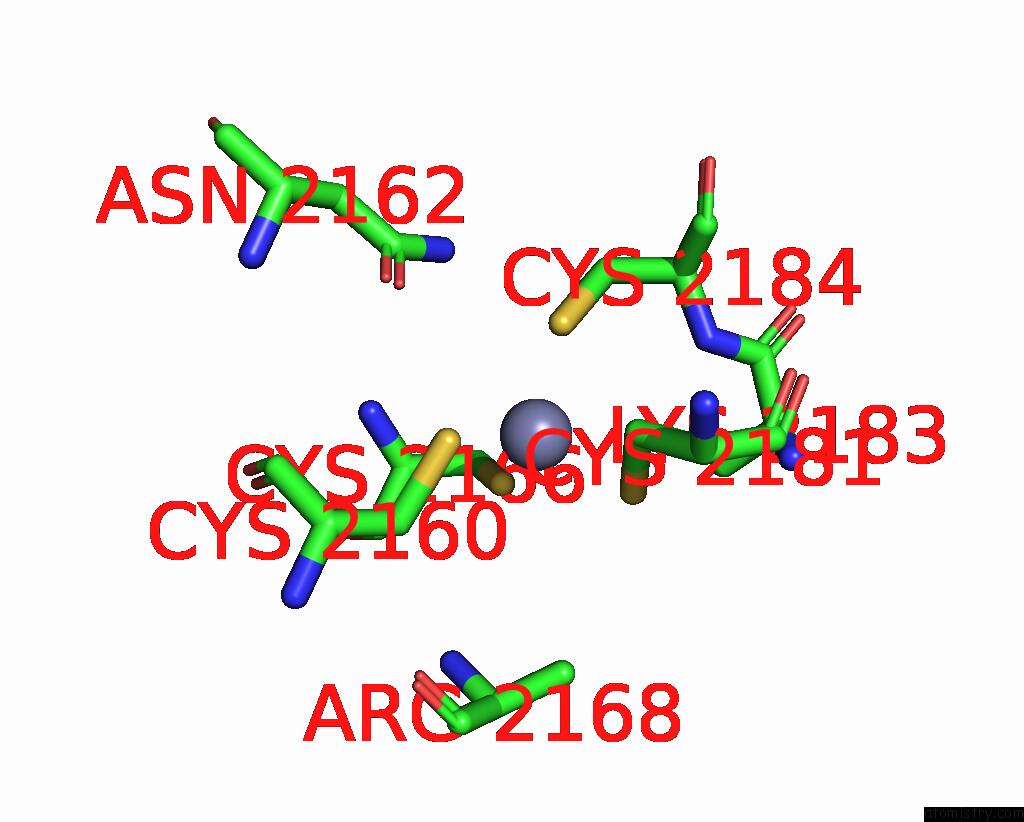
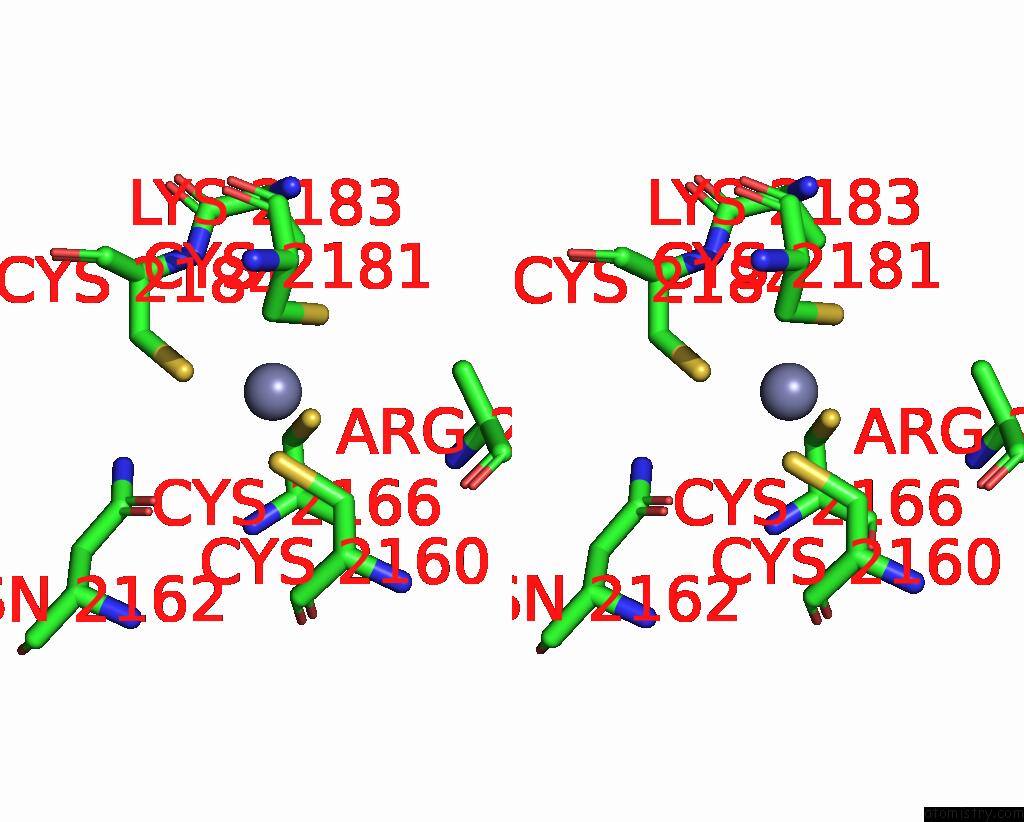
Zinc binding site 4 out of 10 in 8q7h
Go back to
Zinc binding site 4 out
of 10 in the Structure of CUL9-RBX1 Ubiquitin E3 Ligase Complex in Unneddylated and Neddylated Conformation - Focused Cullin Dimer
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 4 of Structure of CUL9-RBX1 Ubiquitin E3 Ligase Complex in Unneddylated and Neddylated Conformation - Focused Cullin Dimer within 5.0Å range:
|
Zinc binding site 5 out of 10 in 8q7h
Go back to
Zinc binding site 5 out
of 10 in the Structure of CUL9-RBX1 Ubiquitin E3 Ligase Complex in Unneddylated and Neddylated Conformation - Focused Cullin Dimer
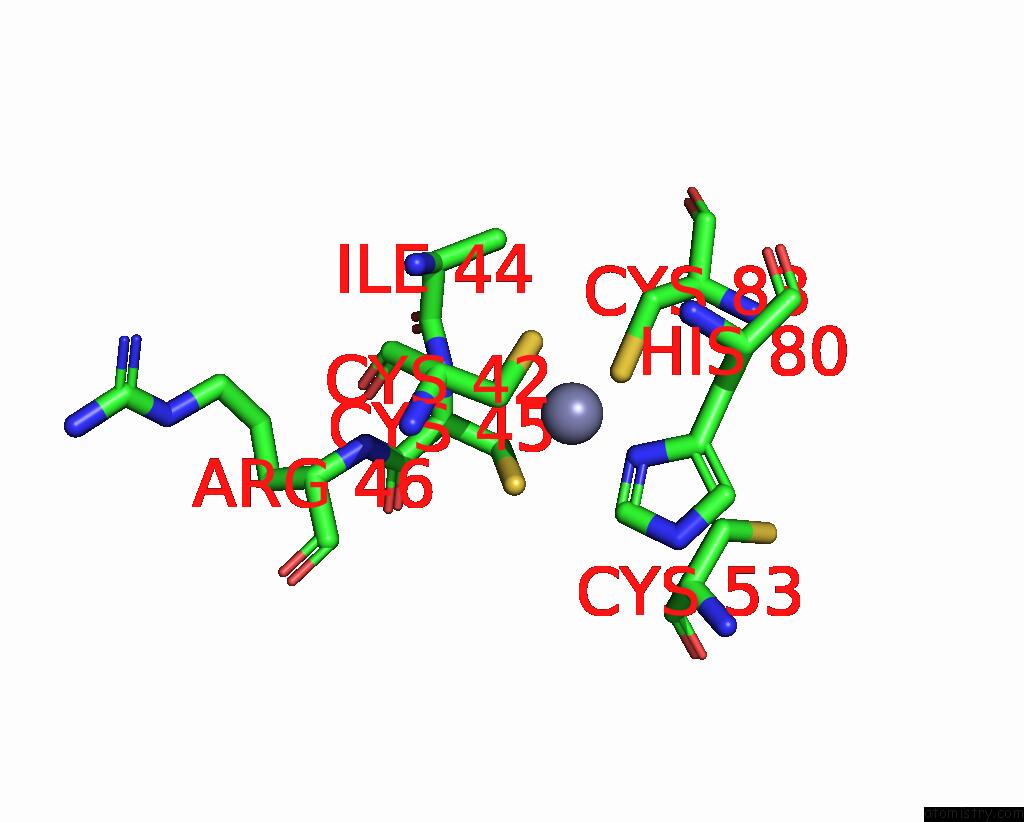
Mono view
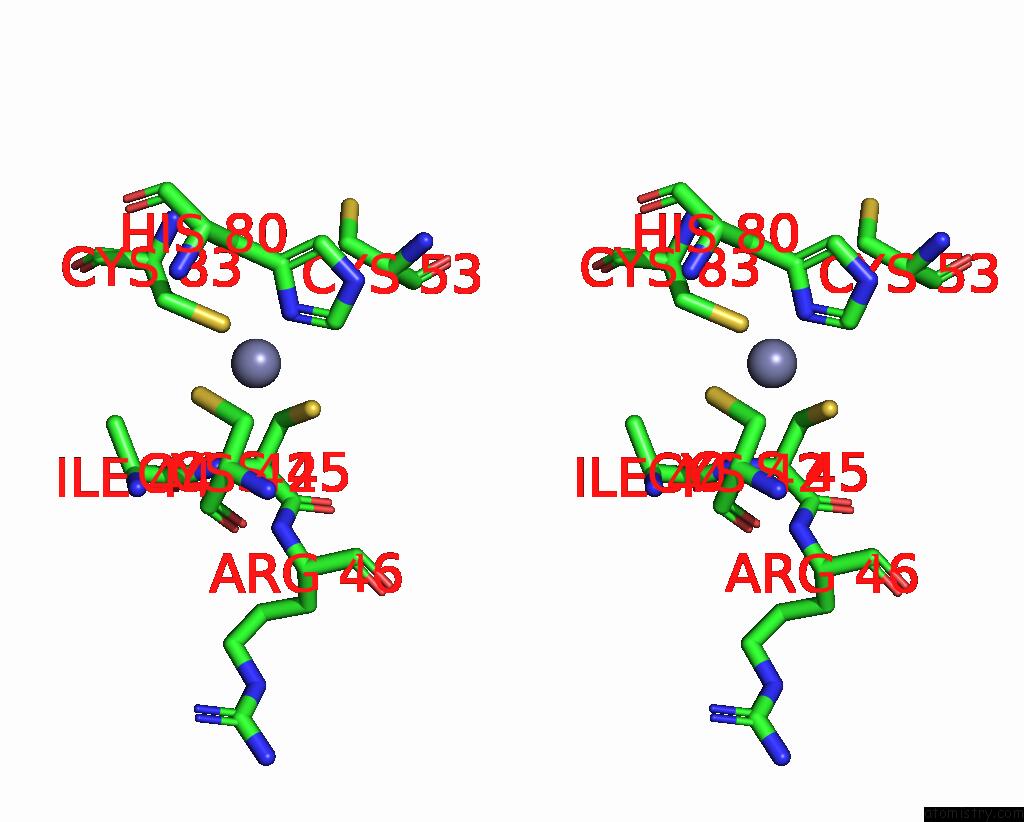
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 5 of Structure of CUL9-RBX1 Ubiquitin E3 Ligase Complex in Unneddylated and Neddylated Conformation - Focused Cullin Dimer within 5.0Å range:
|
Zinc binding site 6 out of 10 in 8q7h
Go back to
Zinc binding site 6 out
of 10 in the Structure of CUL9-RBX1 Ubiquitin E3 Ligase Complex in Unneddylated and Neddylated Conformation - Focused Cullin Dimer
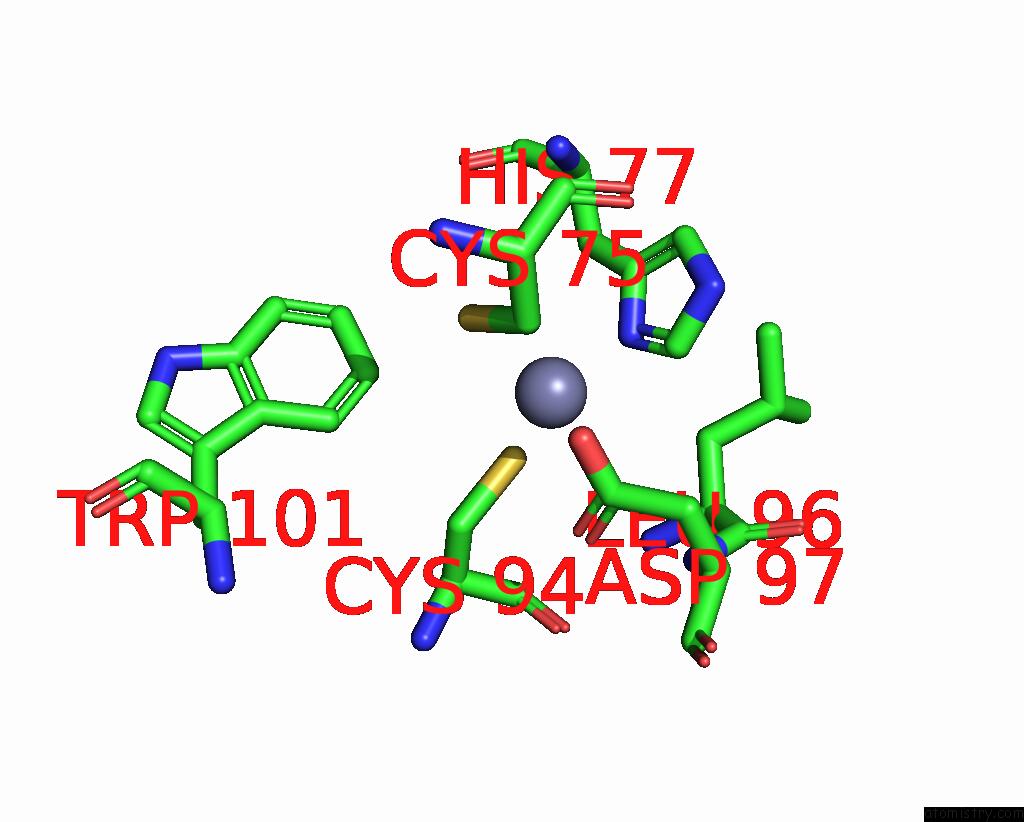
Mono view
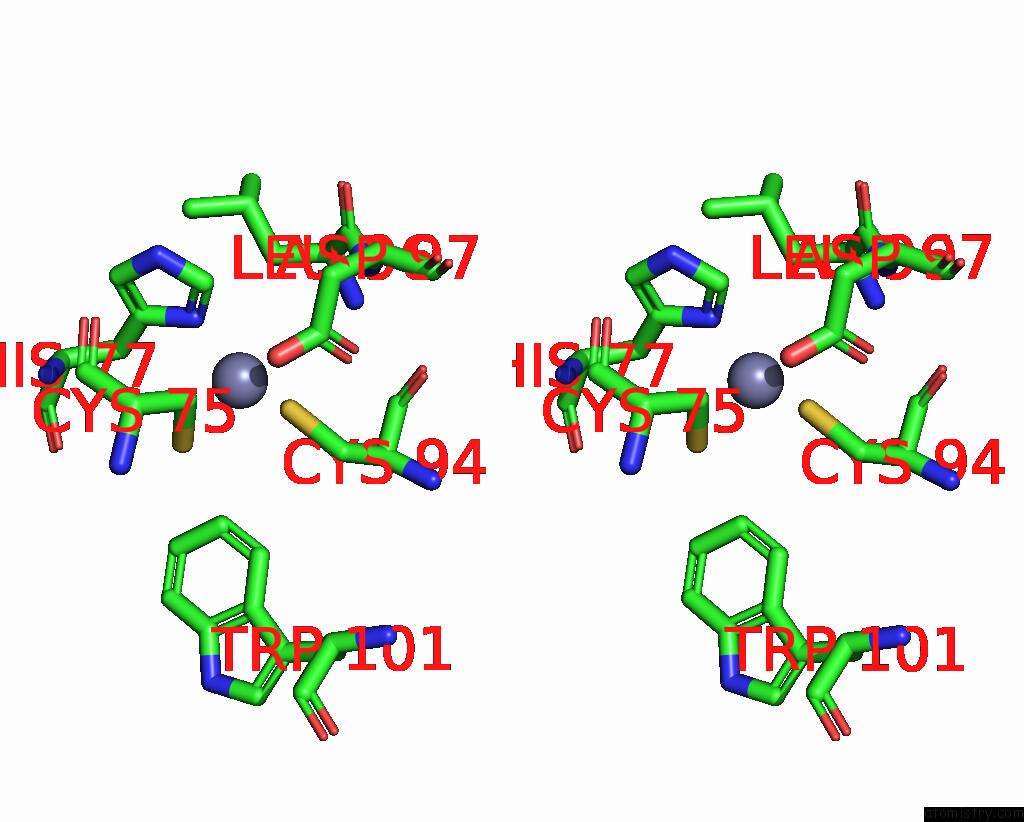
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 6 of Structure of CUL9-RBX1 Ubiquitin E3 Ligase Complex in Unneddylated and Neddylated Conformation - Focused Cullin Dimer within 5.0Å range:
|
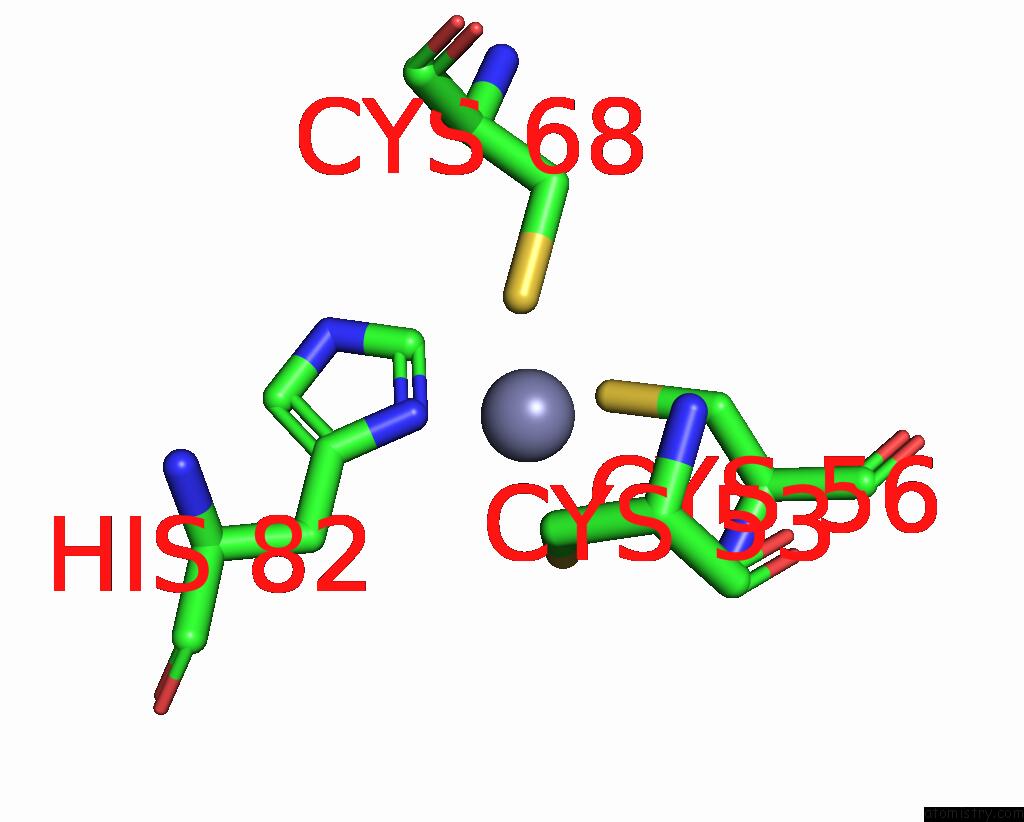
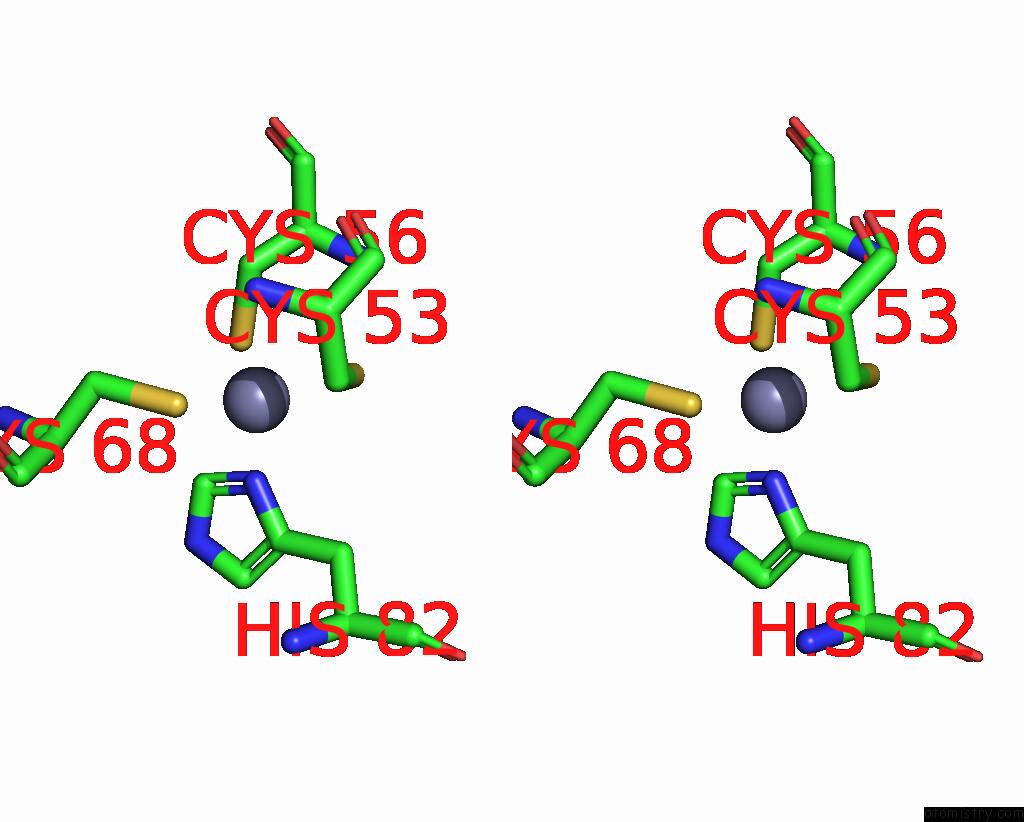
Zinc binding site 7 out of 10 in 8q7h
Go back to
Zinc binding site 7 out
of 10 in the Structure of CUL9-RBX1 Ubiquitin E3 Ligase Complex in Unneddylated and Neddylated Conformation - Focused Cullin Dimer
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 7 of Structure of CUL9-RBX1 Ubiquitin E3 Ligase Complex in Unneddylated and Neddylated Conformation - Focused Cullin Dimer within 5.0Å range:
|
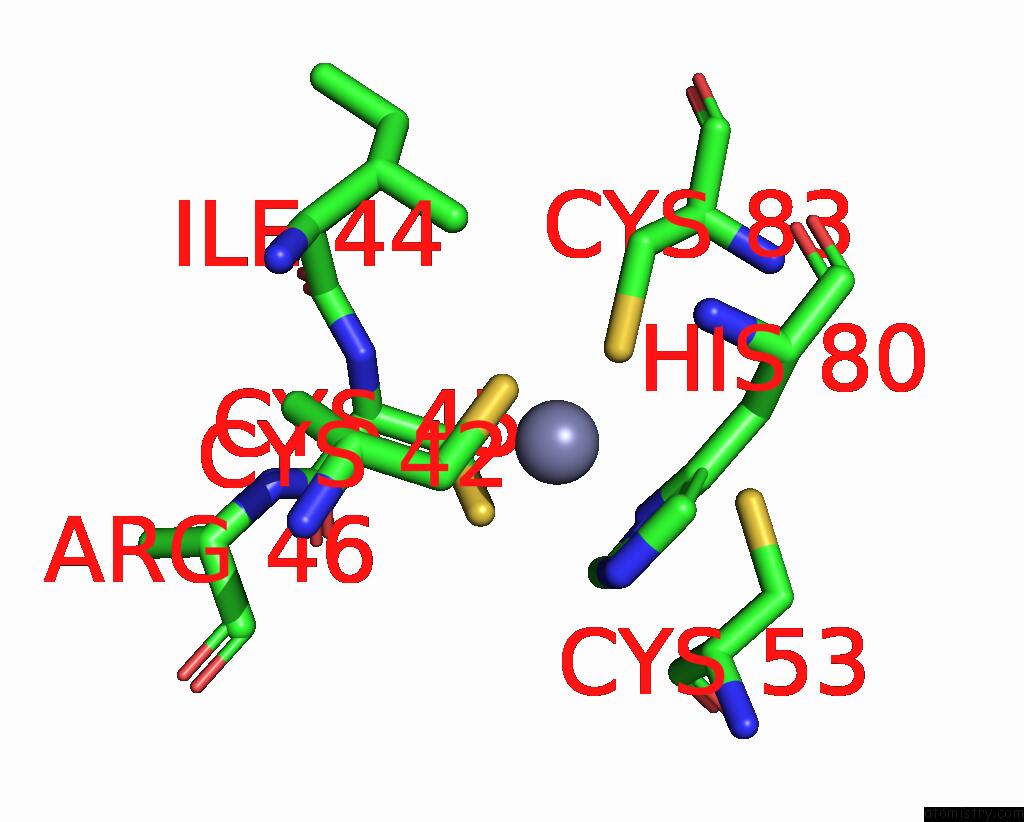
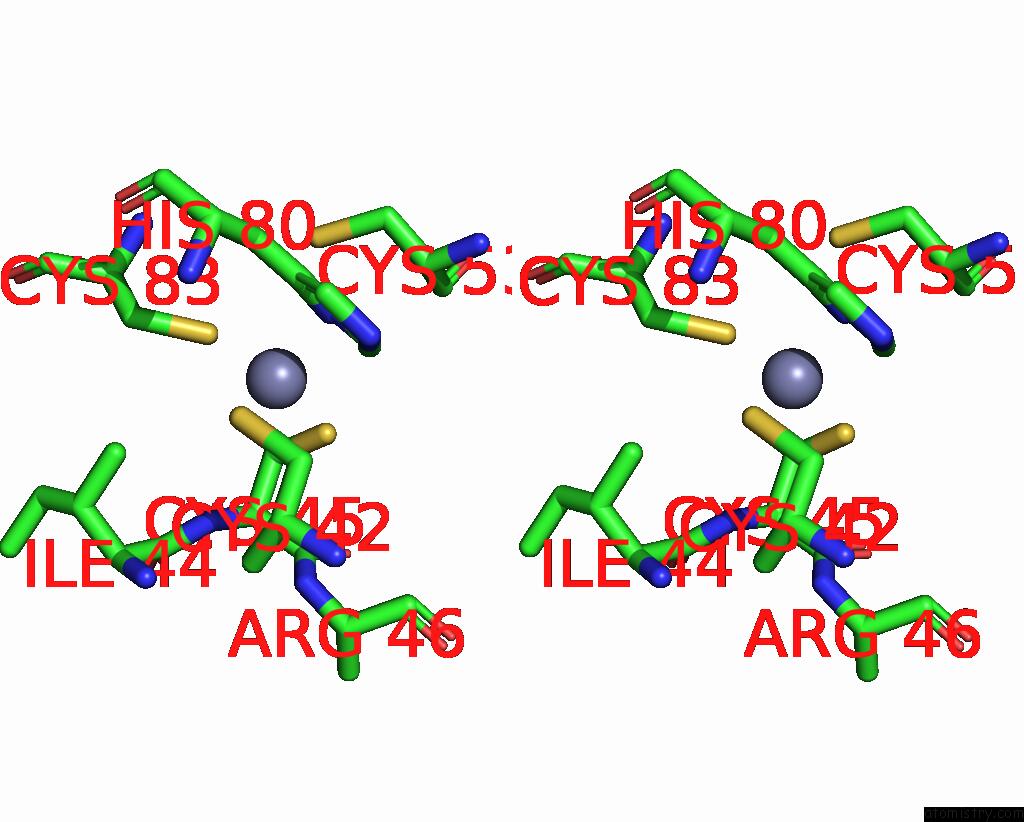
Zinc binding site 8 out of 10 in 8q7h
Go back to
Zinc binding site 8 out
of 10 in the Structure of CUL9-RBX1 Ubiquitin E3 Ligase Complex in Unneddylated and Neddylated Conformation - Focused Cullin Dimer
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 8 of Structure of CUL9-RBX1 Ubiquitin E3 Ligase Complex in Unneddylated and Neddylated Conformation - Focused Cullin Dimer within 5.0Å range:
|
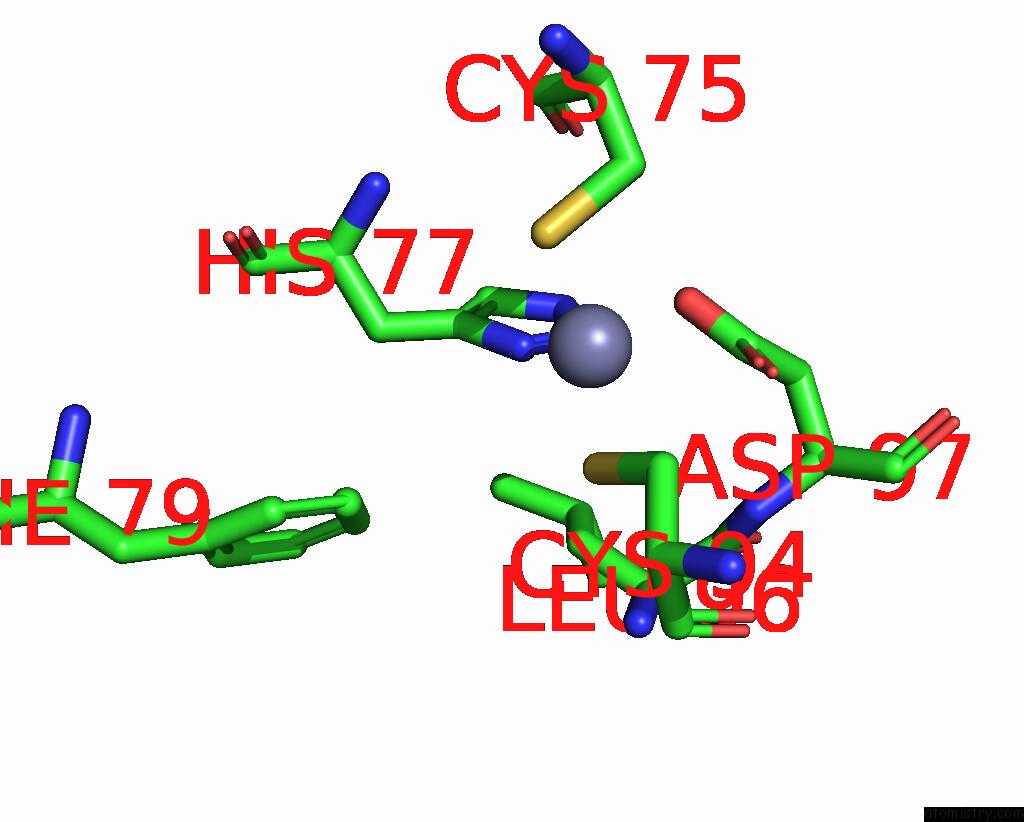
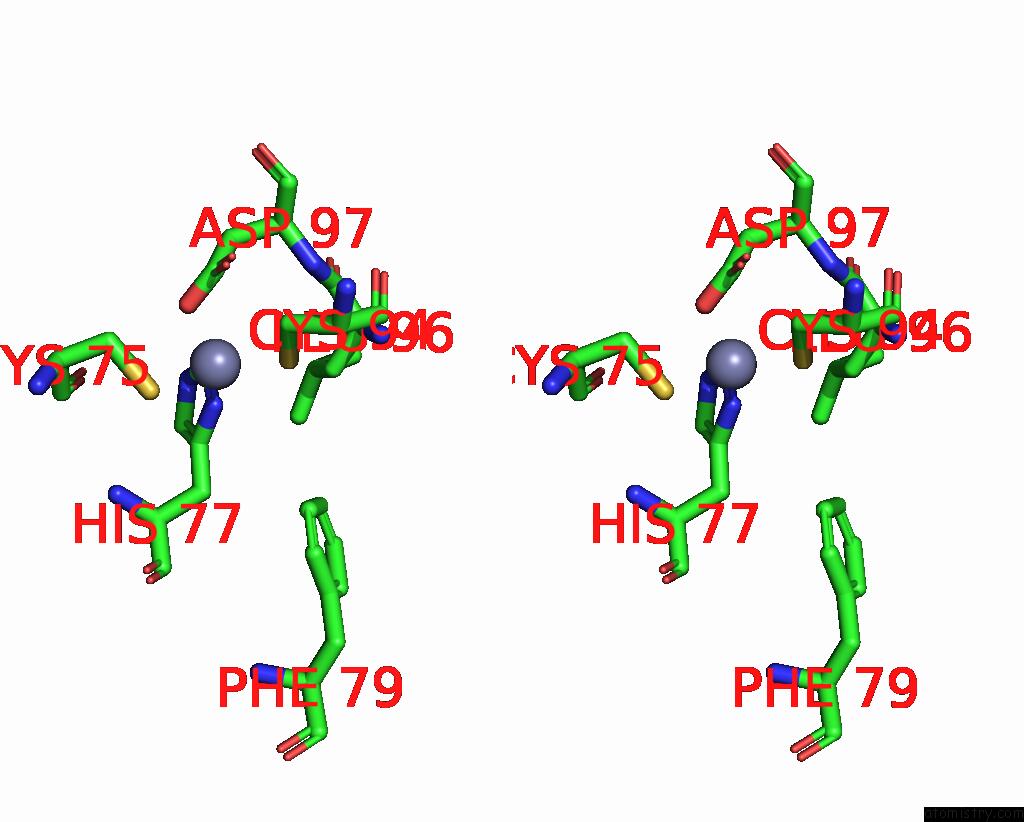
Zinc binding site 9 out of 10 in 8q7h
Go back to
Zinc binding site 9 out
of 10 in the Structure of CUL9-RBX1 Ubiquitin E3 Ligase Complex in Unneddylated and Neddylated Conformation - Focused Cullin Dimer
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 9 of Structure of CUL9-RBX1 Ubiquitin E3 Ligase Complex in Unneddylated and Neddylated Conformation - Focused Cullin Dimer within 5.0Å range:
|
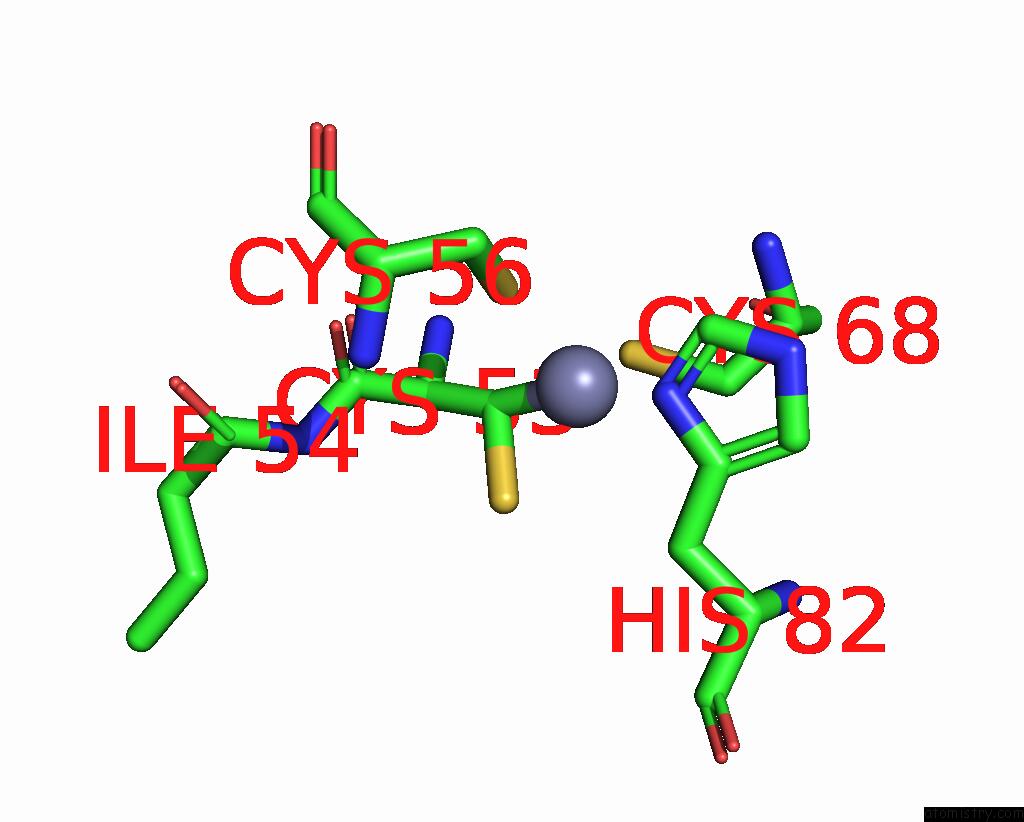
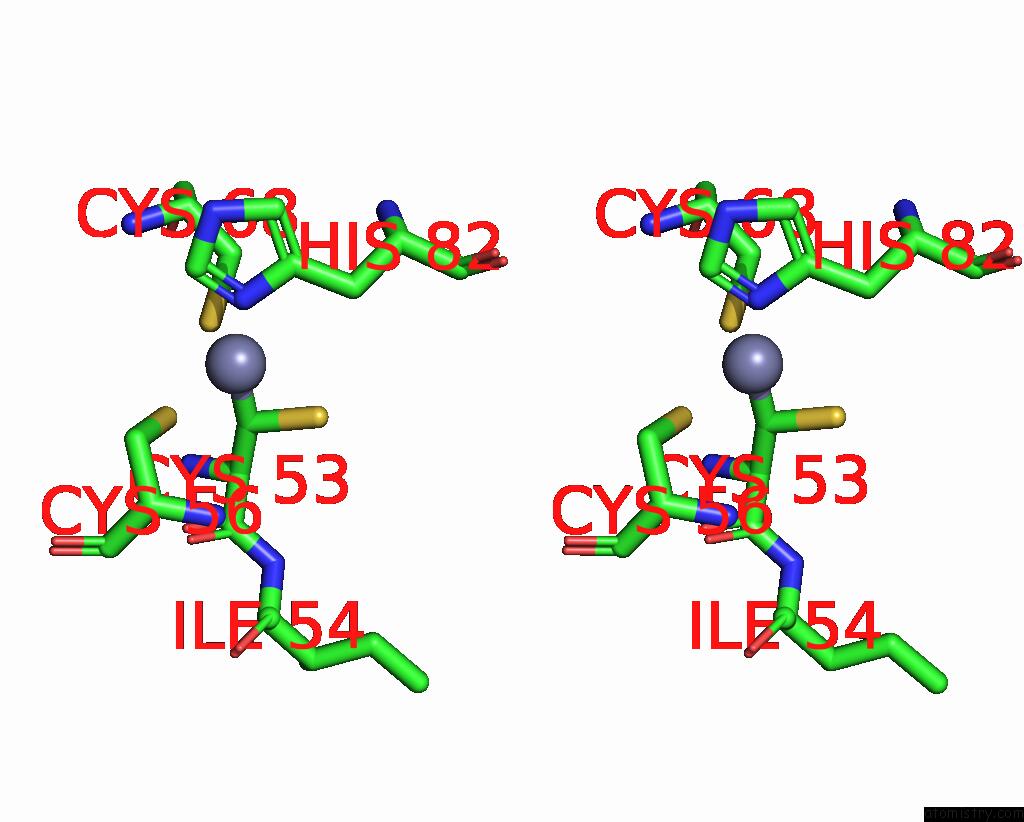
Zinc binding site 10 out of 10 in 8q7h
Go back to
Zinc binding site 10 out
of 10 in the Structure of CUL9-RBX1 Ubiquitin E3 Ligase Complex in Unneddylated and Neddylated Conformation - Focused Cullin Dimer
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 10 of Structure of CUL9-RBX1 Ubiquitin E3 Ligase Complex in Unneddylated and Neddylated Conformation - Focused Cullin Dimer within 5.0Å range:
|
Reference:
L.V.M.Hopf,
D.Horn-Ghetko,
B.A.Schulman.
Noncanonical Assembly, Neddylation and Chimeric Cullin Ring/Rbr Ubiquitylation By the 1.8 Mda CUL9 E3 Ligase Complex Nat.Struct.Mol.Biol. 2024.
ISSN: ESSN 1545-9985
DOI: 10.1038/S41594-024-01257-Y
Page generated: Fri Aug 22 12:34:39 2025
ISSN: ESSN 1545-9985
DOI: 10.1038/S41594-024-01257-Y
Last articles
Zn in 9DBYZn in 9DBF
Zn in 9DAZ
Zn in 9DBD
Zn in 9D89
Zn in 9DAK
Zn in 9D5J
Zn in 9CXJ
Zn in 9CXI
Zn in 9D60