Zinc »
PDB 8e3e-8ebv »
8e40 »
Zinc in PDB 8e40: Full-Length APOBEC3G in Complex with Hiv-1 Vif, Cbf-Beta, and Fork Rna
Enzymatic activity of Full-Length APOBEC3G in Complex with Hiv-1 Vif, Cbf-Beta, and Fork Rna
All present enzymatic activity of Full-Length APOBEC3G in Complex with Hiv-1 Vif, Cbf-Beta, and Fork Rna:
3.5.4.38;
3.5.4.38;
Zinc Binding Sites:
The binding sites of Zinc atom in the Full-Length APOBEC3G in Complex with Hiv-1 Vif, Cbf-Beta, and Fork Rna
(pdb code 8e40). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 2 binding sites of Zinc where determined in the Full-Length APOBEC3G in Complex with Hiv-1 Vif, Cbf-Beta, and Fork Rna, PDB code: 8e40:
Jump to Zinc binding site number: 1; 2;
In total 2 binding sites of Zinc where determined in the Full-Length APOBEC3G in Complex with Hiv-1 Vif, Cbf-Beta, and Fork Rna, PDB code: 8e40:
Jump to Zinc binding site number: 1; 2;
Zinc binding site 1 out of 2 in 8e40
Go back to
Zinc binding site 1 out
of 2 in the Full-Length APOBEC3G in Complex with Hiv-1 Vif, Cbf-Beta, and Fork Rna
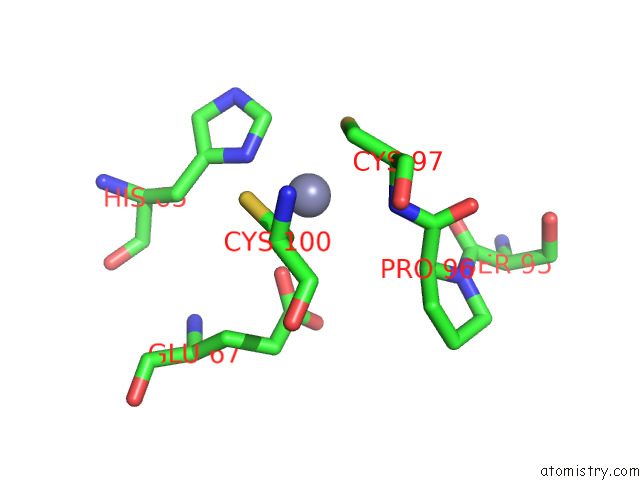
Mono view
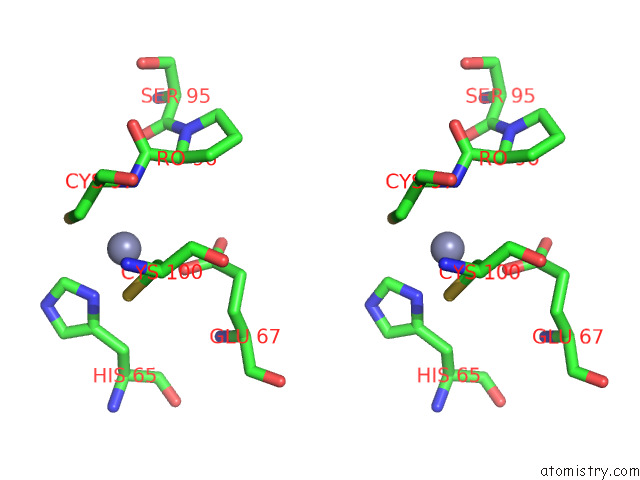
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Full-Length APOBEC3G in Complex with Hiv-1 Vif, Cbf-Beta, and Fork Rna within 5.0Å range:
|
Zinc binding site 2 out of 2 in 8e40
Go back to
Zinc binding site 2 out
of 2 in the Full-Length APOBEC3G in Complex with Hiv-1 Vif, Cbf-Beta, and Fork Rna
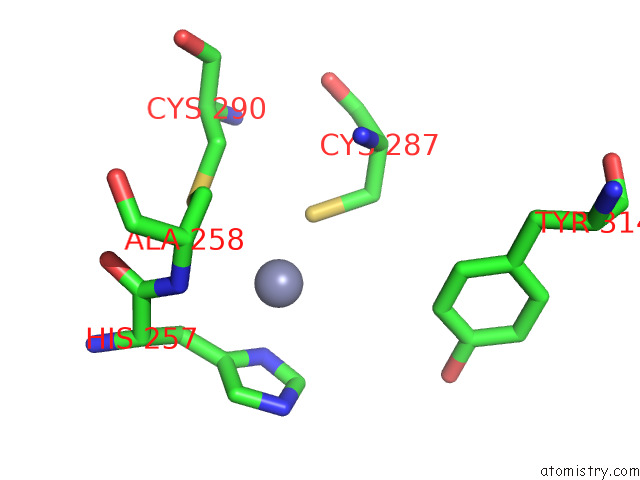
Mono view
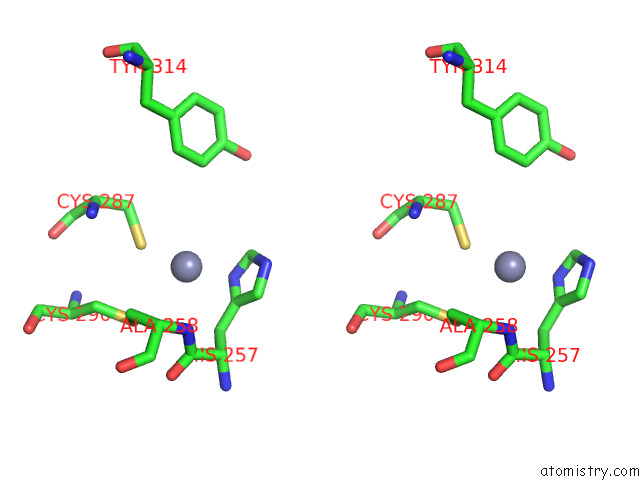
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of Full-Length APOBEC3G in Complex with Hiv-1 Vif, Cbf-Beta, and Fork Rna within 5.0Å range:
|
Reference:
F.Ito,
A.L.Alvarez-Cabrera,
S.Liu,
H.Yang,
A.Shiriaeva,
Z.H.Zhou,
X.S.Chen.
Structural Basis For Hiv-1 Antagonism of Host APOBEC3G Via Cullin E3 Ligase. Sci Adv V. 9 E3168 2023.
ISSN: ESSN 2375-2548
PubMed: 36598981
DOI: 10.1126/SCIADV.ADE3168
Page generated: Fri Aug 22 09:24:45 2025
ISSN: ESSN 2375-2548
PubMed: 36598981
DOI: 10.1126/SCIADV.ADE3168
Last articles
Zn in 8S8GZn in 8S8H
Zn in 8S8F
Zn in 8S8D
Zn in 8S8E
Zn in 8S5N
Zn in 8S7V
Zn in 8S52
Zn in 8S55
Zn in 8S4F