Zinc »
PDB 8e3e-8ebv »
8e3s »
Zinc in PDB 8e3s: Cryoem Structure of Yeast Arginyltransferase 1 (ATE1)
Enzymatic activity of Cryoem Structure of Yeast Arginyltransferase 1 (ATE1)
All present enzymatic activity of Cryoem Structure of Yeast Arginyltransferase 1 (ATE1):
2.3.2.8;
2.3.2.8;
Zinc Binding Sites:
The binding sites of Zinc atom in the Cryoem Structure of Yeast Arginyltransferase 1 (ATE1)
(pdb code 8e3s). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total only one binding site of Zinc was determined in the Cryoem Structure of Yeast Arginyltransferase 1 (ATE1), PDB code: 8e3s:
In total only one binding site of Zinc was determined in the Cryoem Structure of Yeast Arginyltransferase 1 (ATE1), PDB code: 8e3s:
Zinc binding site 1 out of 1 in 8e3s
Go back to
Zinc binding site 1 out
of 1 in the Cryoem Structure of Yeast Arginyltransferase 1 (ATE1)
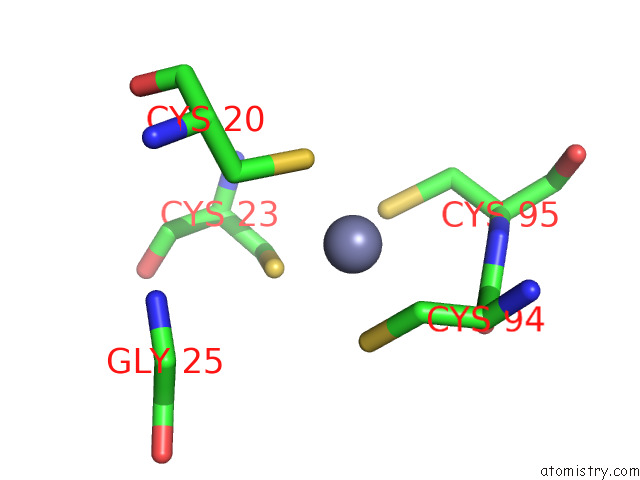
Mono view
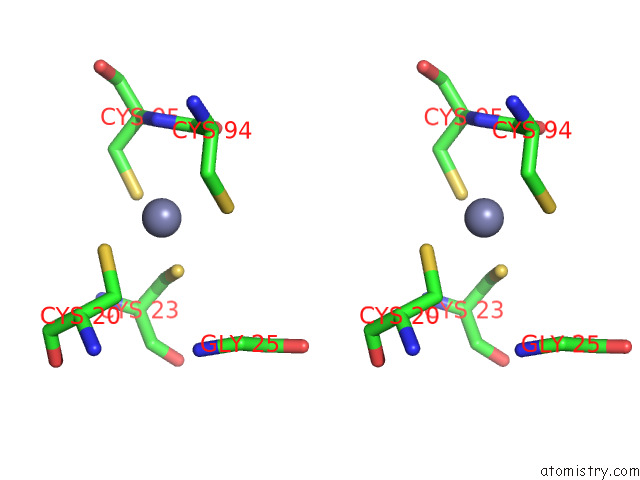
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Cryoem Structure of Yeast Arginyltransferase 1 (ATE1) within 5.0Å range:
|
Reference:
T.Abeywansha,
W.Huang,
X.Ye,
A.Nawrocki,
X.Lan,
E.Jankowsky,
D.J.Taylor,
Y.Zhang.
The Structural Basis of Trna Recognition By Arginyl-Trna-Protein Transferase Nat Commun V. 14 2232 2023.
ISSN: ESSN 2041-1723
DOI: 10.1038/S41467-023-38004-8
Page generated: Fri Aug 22 09:24:31 2025
ISSN: ESSN 2041-1723
DOI: 10.1038/S41467-023-38004-8
Last articles
Zn in 8SD0Zn in 8S9R
Zn in 8SCZ
Zn in 8S93
Zn in 8SAG
Zn in 8SAF
Zn in 8S8K
Zn in 8S8J
Zn in 8S8X
Zn in 8S8W