Zinc in PDB 7zxe: Structure of Snapc Containing Pol II Pre-Initiation Complex Bound to U1 Snrna Promoter (Oc)
Enzymatic activity of Structure of Snapc Containing Pol II Pre-Initiation Complex Bound to U1 Snrna Promoter (Oc)
All present enzymatic activity of Structure of Snapc Containing Pol II Pre-Initiation Complex Bound to U1 Snrna Promoter (Oc):
2.3.1.48;
2.3.1.48;
Zinc Binding Sites:
The binding sites of Zinc atom in the Structure of Snapc Containing Pol II Pre-Initiation Complex Bound to U1 Snrna Promoter (Oc)
(pdb code 7zxe). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 2 binding sites of Zinc where determined in the Structure of Snapc Containing Pol II Pre-Initiation Complex Bound to U1 Snrna Promoter (Oc), PDB code: 7zxe:
Jump to Zinc binding site number: 1; 2;
In total 2 binding sites of Zinc where determined in the Structure of Snapc Containing Pol II Pre-Initiation Complex Bound to U1 Snrna Promoter (Oc), PDB code: 7zxe:
Jump to Zinc binding site number: 1; 2;
Zinc binding site 1 out of 2 in 7zxe
Go back to
Zinc binding site 1 out
of 2 in the Structure of Snapc Containing Pol II Pre-Initiation Complex Bound to U1 Snrna Promoter (Oc)
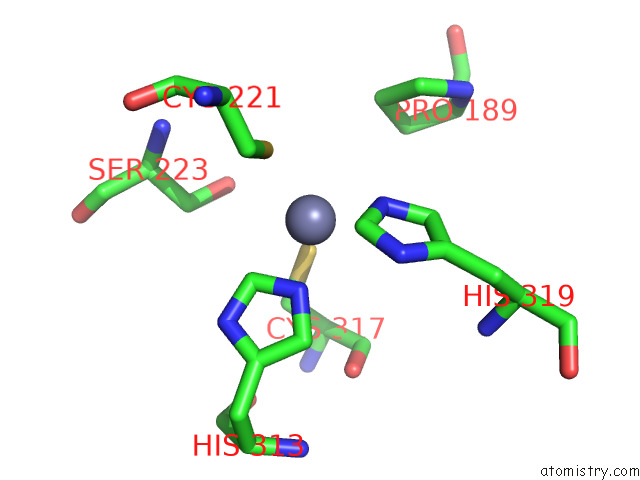
Mono view
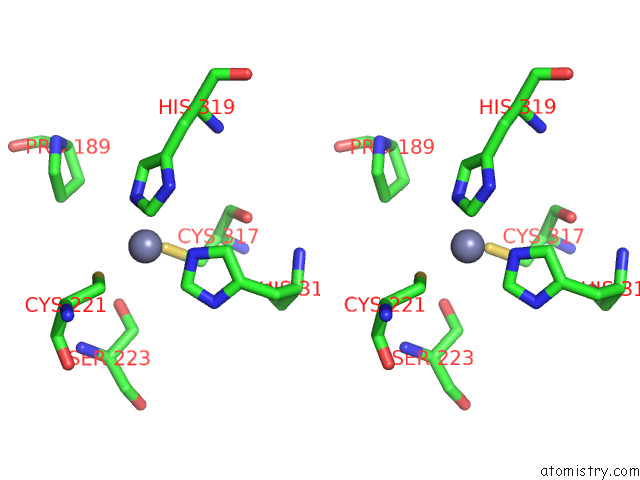
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Structure of Snapc Containing Pol II Pre-Initiation Complex Bound to U1 Snrna Promoter (Oc) within 5.0Å range:
|
Zinc binding site 2 out of 2 in 7zxe
Go back to
Zinc binding site 2 out
of 2 in the Structure of Snapc Containing Pol II Pre-Initiation Complex Bound to U1 Snrna Promoter (Oc)
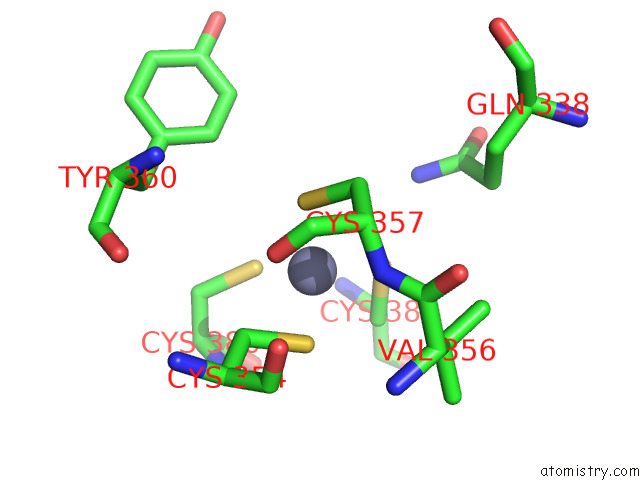
Mono view
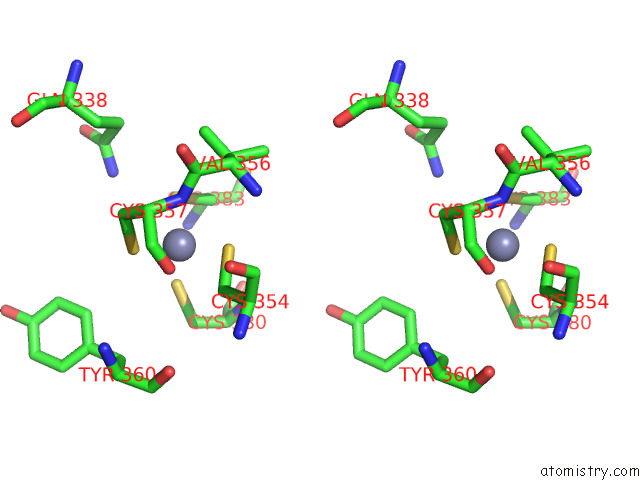
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of Structure of Snapc Containing Pol II Pre-Initiation Complex Bound to U1 Snrna Promoter (Oc) within 5.0Å range:
|
Reference:
S.Rengachari,
S.Schilbach,
T.Kaliyappan,
J.Gouge,
K.Zumer,
J.Schwarz,
H.Urlaub,
C.Dienemann,
A.Vannini,
P.Cramer.
Structural Basis of Snapc-Dependent Snrna Transcription Initiation By Rna Polymerase II. Nat.Struct.Mol.Biol. V. 29 1159 2022.
ISSN: ESSN 1545-9985
PubMed: 36424526
DOI: 10.1038/S41594-022-00857-W
Page generated: Wed Oct 30 17:21:23 2024
ISSN: ESSN 1545-9985
PubMed: 36424526
DOI: 10.1038/S41594-022-00857-W
Last articles
Zn in 9MJ5Zn in 9HNW
Zn in 9G0L
Zn in 9FNE
Zn in 9DZN
Zn in 9E0I
Zn in 9D32
Zn in 9DAK
Zn in 8ZXC
Zn in 8ZUF