Zinc »
PDB 7ysf-7z7e »
7z71 »
Zinc in PDB 7z71: Crystal Structure of P63 Dbd in Complex with Darpin C14
Protein crystallography data
The structure of Crystal Structure of P63 Dbd in Complex with Darpin C14, PDB code: 7z71
was solved by
A.Chaikuad,
A.Strubel,
V.Doetsch,
S.Knapp,
Structural Genomics Consortium(Sgc),
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 42.69 / 1.85 |
| Space group | P 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 53.033, 63.93, 65.504, 114.46, 94.6, 104.06 |
| R / Rfree (%) | 14.2 / 18.1 |
Zinc Binding Sites:
The binding sites of Zinc atom in the Crystal Structure of P63 Dbd in Complex with Darpin C14
(pdb code 7z71). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 2 binding sites of Zinc where determined in the Crystal Structure of P63 Dbd in Complex with Darpin C14, PDB code: 7z71:
Jump to Zinc binding site number: 1; 2;
In total 2 binding sites of Zinc where determined in the Crystal Structure of P63 Dbd in Complex with Darpin C14, PDB code: 7z71:
Jump to Zinc binding site number: 1; 2;
Zinc binding site 1 out of 2 in 7z71
Go back to
Zinc binding site 1 out
of 2 in the Crystal Structure of P63 Dbd in Complex with Darpin C14
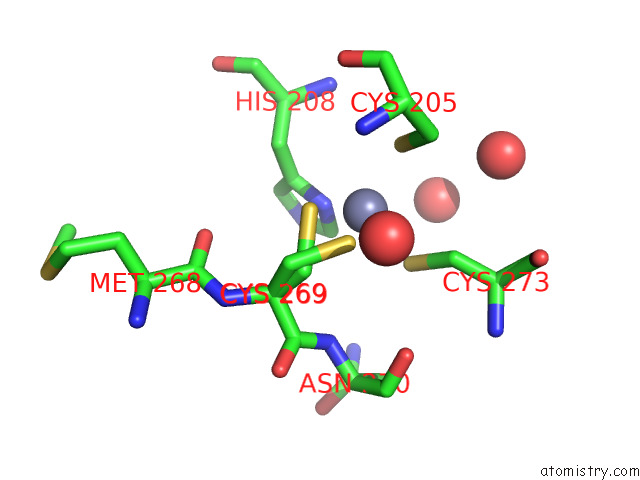
Mono view
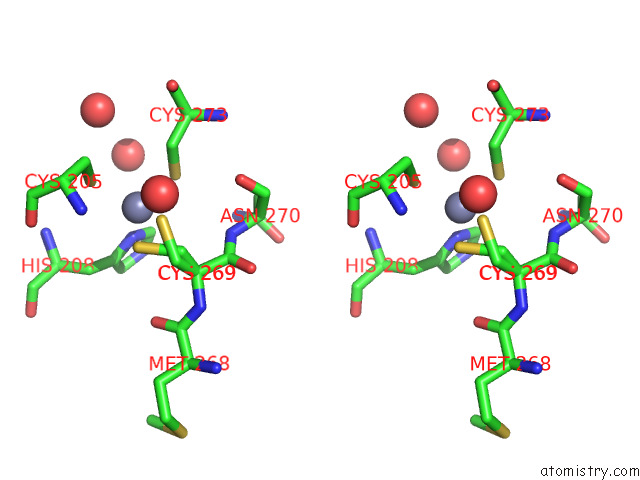
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Crystal Structure of P63 Dbd in Complex with Darpin C14 within 5.0Å range:
|
Zinc binding site 2 out of 2 in 7z71
Go back to
Zinc binding site 2 out
of 2 in the Crystal Structure of P63 Dbd in Complex with Darpin C14
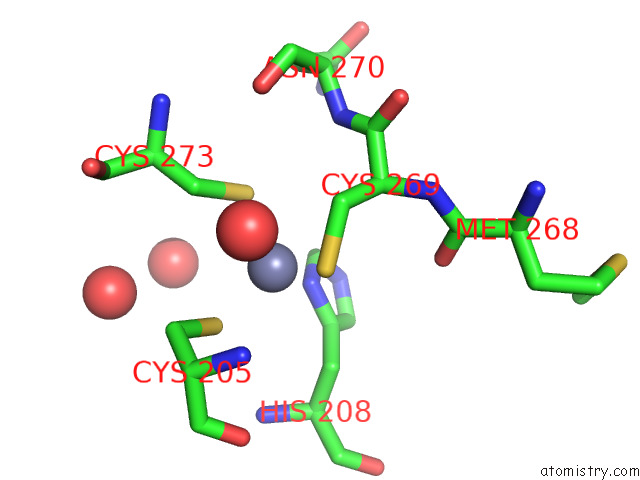
Mono view
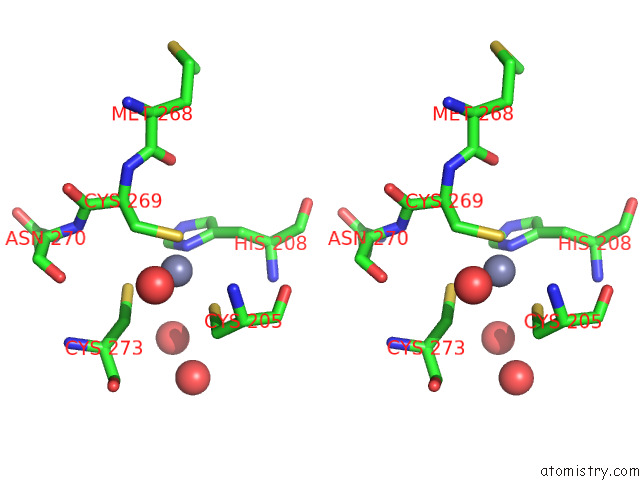
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of Crystal Structure of P63 Dbd in Complex with Darpin C14 within 5.0Å range:
|
Reference:
A.Strubel,
P.Munick,
A.Chaikuad,
B.Dreier,
J.Schaefer,
J.Gebel,
C.Osterburg,
M.Tuppi,
B.Schafer,
S.Knapp,
A.Pluckthun,
V.Dotsch.
Designed Ankyrin Repeat Proteins As A Tool Box For Analyzing P63. Cell Death Differ. V. 29 2445 2022.
ISSN: ISSN 1350-9047
PubMed: 35717504
DOI: 10.1038/S41418-022-01030-Y
Page generated: Fri Aug 22 07:23:56 2025
ISSN: ISSN 1350-9047
PubMed: 35717504
DOI: 10.1038/S41418-022-01030-Y
Last articles
Zn in 8X4LZn in 8X5J
Zn in 8X6M
Zn in 8X4P
Zn in 8X4O
Zn in 8X46
Zn in 8X4I
Zn in 8X41
Zn in 8X47
Zn in 8X3W