Zinc »
PDB 7m5o-7ml4 »
7m8e »
Zinc in PDB 7m8e: E.Coli Rnap-Rapa Elongation Complex
Enzymatic activity of E.Coli Rnap-Rapa Elongation Complex
All present enzymatic activity of E.Coli Rnap-Rapa Elongation Complex:
2.7.7.6;
2.7.7.6;
Other elements in 7m8e:
The structure of E.Coli Rnap-Rapa Elongation Complex also contains other interesting chemical elements:
| Magnesium | (Mg) | 1 atom |
Zinc Binding Sites:
The binding sites of Zinc atom in the E.Coli Rnap-Rapa Elongation Complex
(pdb code 7m8e). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 2 binding sites of Zinc where determined in the E.Coli Rnap-Rapa Elongation Complex, PDB code: 7m8e:
Jump to Zinc binding site number: 1; 2;
In total 2 binding sites of Zinc where determined in the E.Coli Rnap-Rapa Elongation Complex, PDB code: 7m8e:
Jump to Zinc binding site number: 1; 2;
Zinc binding site 1 out of 2 in 7m8e
Go back to
Zinc binding site 1 out
of 2 in the E.Coli Rnap-Rapa Elongation Complex
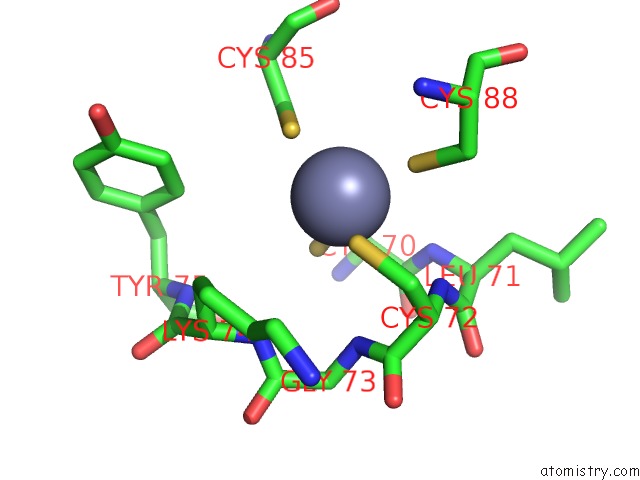
Mono view
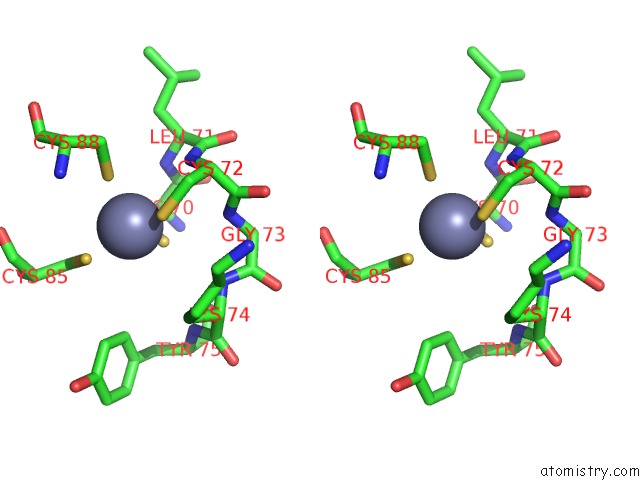
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of E.Coli Rnap-Rapa Elongation Complex within 5.0Å range:
|
Zinc binding site 2 out of 2 in 7m8e
Go back to
Zinc binding site 2 out
of 2 in the E.Coli Rnap-Rapa Elongation Complex
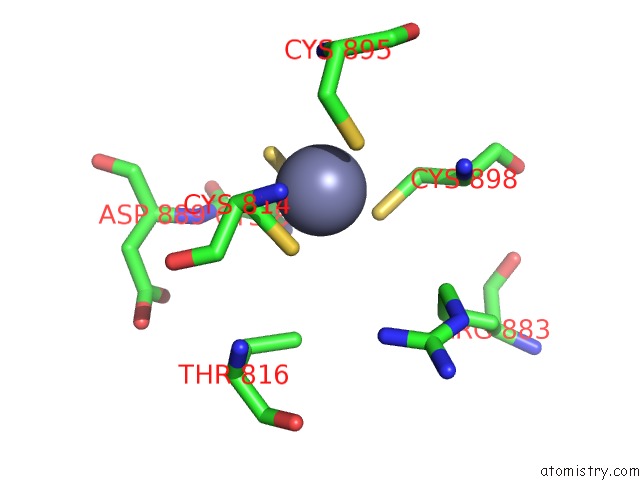
Mono view
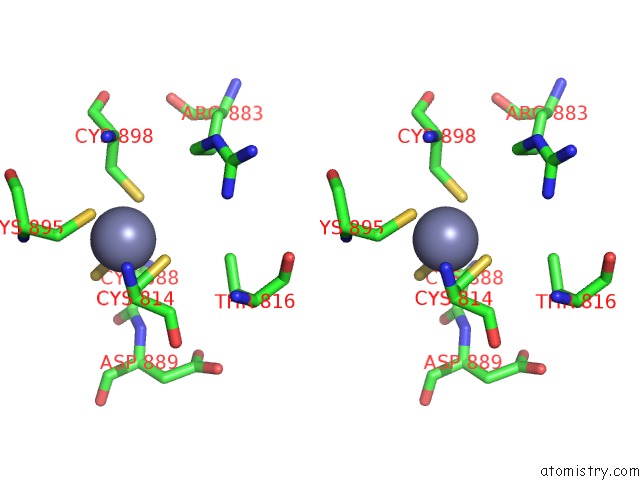
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of E.Coli Rnap-Rapa Elongation Complex within 5.0Å range:
|
Reference:
W.Shi,
W.Zhou,
M.Chen,
Y.Yang,
Y.Hu,
B.Liu.
Structural Basis For Activation of SWI2/SNF2 Atpase Rapa By Rna Polymerase Nucleic Acids Res. 2021.
ISSN: ESSN 1362-4962
Page generated: Fri Aug 22 02:04:38 2025
ISSN: ESSN 1362-4962
Last articles
Zn in 7X76Zn in 7XAZ
Zn in 7XAB
Zn in 7XAA
Zn in 7XA7
Zn in 7X9X
Zn in 7XA4
Zn in 7X97
Zn in 7X8A
Zn in 7X7R