Zinc »
PDB 6v7a-6vrg »
6vpc »
Zinc in PDB 6vpc: Structure of the SPCAS9 Dna Adenine Base Editor - ABE8E
Other elements in 6vpc:
The structure of Structure of the SPCAS9 Dna Adenine Base Editor - ABE8E also contains other interesting chemical elements:
| Magnesium | (Mg) | 3 atoms |
Zinc Binding Sites:
The binding sites of Zinc atom in the Structure of the SPCAS9 Dna Adenine Base Editor - ABE8E
(pdb code 6vpc). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 2 binding sites of Zinc where determined in the Structure of the SPCAS9 Dna Adenine Base Editor - ABE8E, PDB code: 6vpc:
Jump to Zinc binding site number: 1; 2;
In total 2 binding sites of Zinc where determined in the Structure of the SPCAS9 Dna Adenine Base Editor - ABE8E, PDB code: 6vpc:
Jump to Zinc binding site number: 1; 2;
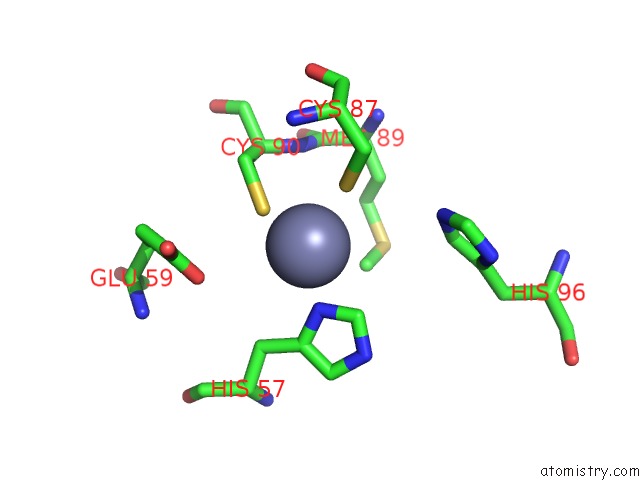
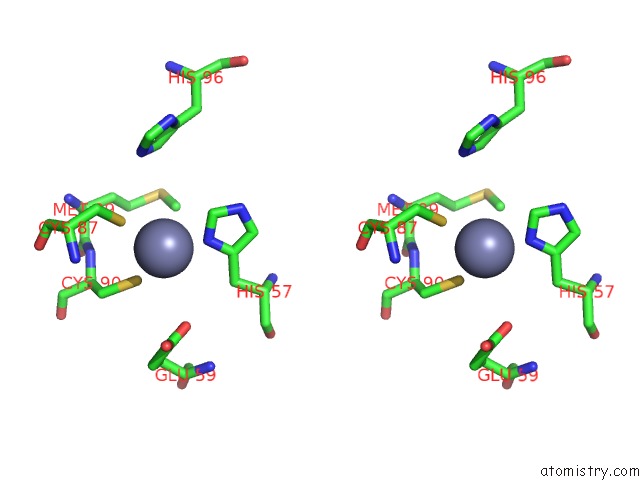
Zinc binding site 1 out of 2 in 6vpc
Go back to
Zinc binding site 1 out
of 2 in the Structure of the SPCAS9 Dna Adenine Base Editor - ABE8E
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Structure of the SPCAS9 Dna Adenine Base Editor - ABE8E within 5.0Å range:
|
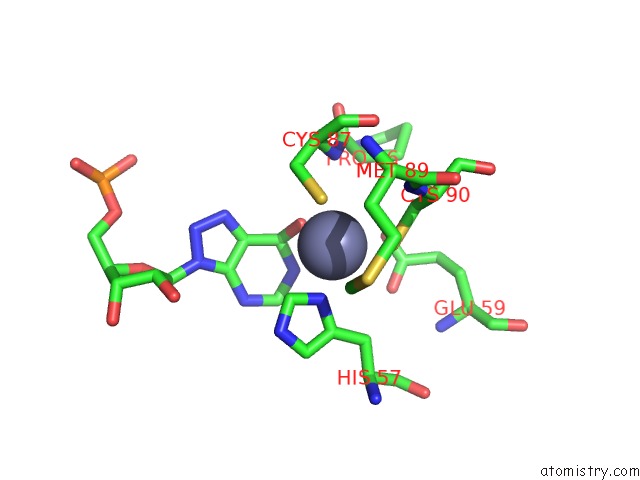
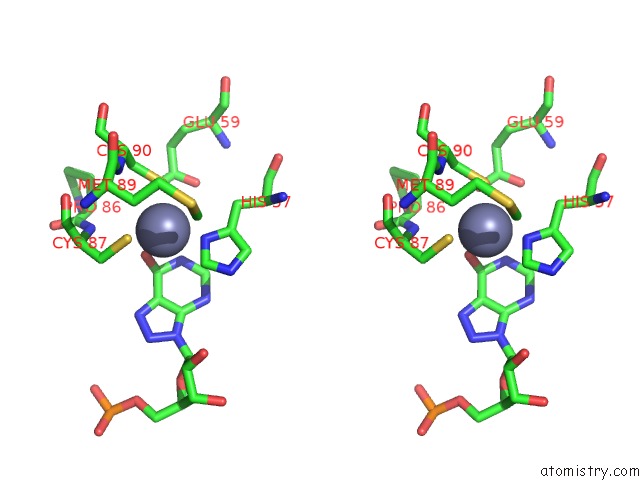
Zinc binding site 2 out of 2 in 6vpc
Go back to
Zinc binding site 2 out
of 2 in the Structure of the SPCAS9 Dna Adenine Base Editor - ABE8E
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of Structure of the SPCAS9 Dna Adenine Base Editor - ABE8E within 5.0Å range:
|
Reference:
A.Lapinaite,
G.J.Knott,
C.M.Palumbo,
E.Lin-Shiao,
M.F.Richter,
K.T.Zhao,
P.A.Beal,
D.R.Liu,
J.A.Doudna.
Dna Capture By A Crispr-CAS9-Guided Adenine Base Editor. Science V. 369 566 2020.
ISSN: ESSN 1095-9203
PubMed: 32732424
DOI: 10.1126/SCIENCE.ABB1390
Page generated: Thu Aug 21 20:34:17 2025
ISSN: ESSN 1095-9203
PubMed: 32732424
DOI: 10.1126/SCIENCE.ABB1390
Last articles
Zn in 7DUGZn in 7DUF
Zn in 7DUB
Zn in 7DUC
Zn in 7DTN
Zn in 7DU2
Zn in 7DTM
Zn in 7DU1
Zn in 7DTE
Zn in 7DTA