Zinc »
PDB 6kam-6km5 »
6kj6 »
Zinc in PDB 6kj6: Cryo-Em Structure of Escherichia Coli Crl Transcription Activation Complex
Enzymatic activity of Cryo-Em Structure of Escherichia Coli Crl Transcription Activation Complex
All present enzymatic activity of Cryo-Em Structure of Escherichia Coli Crl Transcription Activation Complex:
2.7.7.6;
2.7.7.6;
Other elements in 6kj6:
The structure of Cryo-Em Structure of Escherichia Coli Crl Transcription Activation Complex also contains other interesting chemical elements:
| Magnesium | (Mg) | 1 atom |
Zinc Binding Sites:
The binding sites of Zinc atom in the Cryo-Em Structure of Escherichia Coli Crl Transcription Activation Complex
(pdb code 6kj6). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 2 binding sites of Zinc where determined in the Cryo-Em Structure of Escherichia Coli Crl Transcription Activation Complex, PDB code: 6kj6:
Jump to Zinc binding site number: 1; 2;
In total 2 binding sites of Zinc where determined in the Cryo-Em Structure of Escherichia Coli Crl Transcription Activation Complex, PDB code: 6kj6:
Jump to Zinc binding site number: 1; 2;
Zinc binding site 1 out of 2 in 6kj6
Go back to
Zinc binding site 1 out
of 2 in the Cryo-Em Structure of Escherichia Coli Crl Transcription Activation Complex
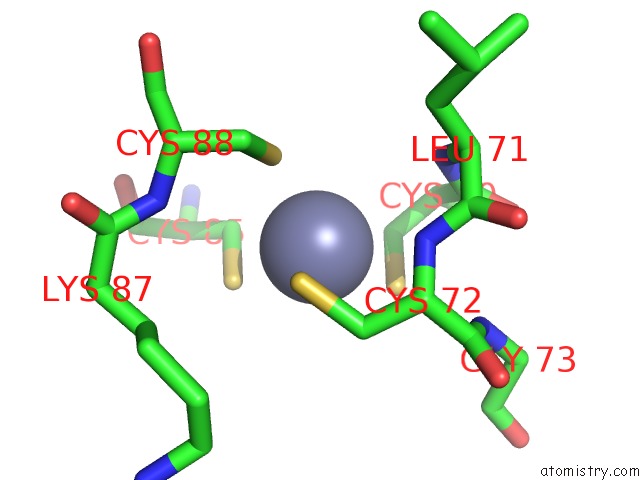
Mono view
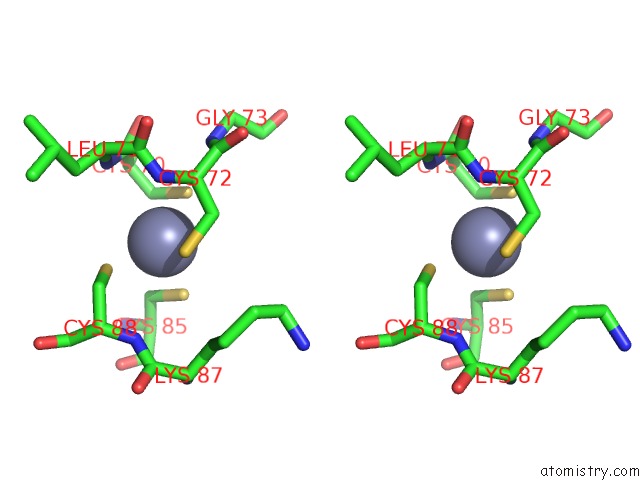
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Cryo-Em Structure of Escherichia Coli Crl Transcription Activation Complex within 5.0Å range:
|
Zinc binding site 2 out of 2 in 6kj6
Go back to
Zinc binding site 2 out
of 2 in the Cryo-Em Structure of Escherichia Coli Crl Transcription Activation Complex
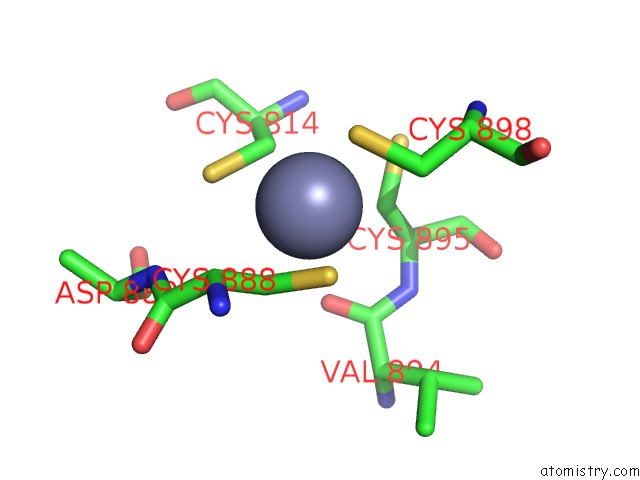
Mono view
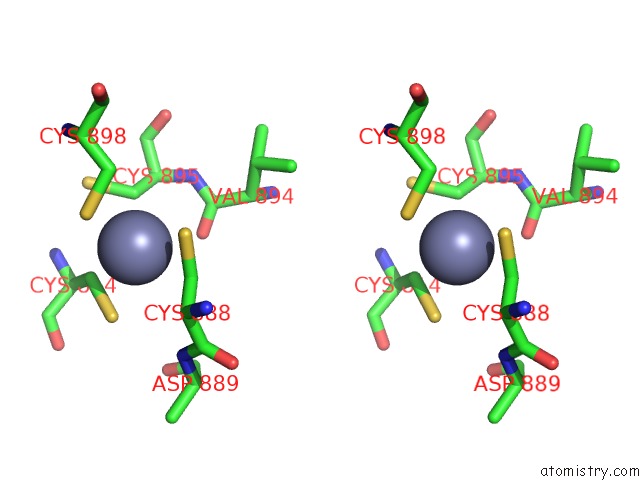
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of Cryo-Em Structure of Escherichia Coli Crl Transcription Activation Complex within 5.0Å range:
|
Reference:
J.Xu,
K.Cui,
L.Shen,
J.Shi,
L.Li,
L.You,
C.Fang,
G.Zhao,
Y.Feng,
B.Yang,
Y.Zhang.
Crl Activates Transcription By Stabilizing Active Conformation of the Master Stress Transcription Initiation Factor. Elife V. 8 2019.
ISSN: ESSN 2050-084X
PubMed: 31846423
DOI: 10.7554/ELIFE.50928
Page generated: Tue Oct 29 01:46:05 2024
ISSN: ESSN 2050-084X
PubMed: 31846423
DOI: 10.7554/ELIFE.50928
Last articles
Zn in 9J0NZn in 9J0O
Zn in 9J0P
Zn in 9FJX
Zn in 9EKB
Zn in 9C0F
Zn in 9CAH
Zn in 9CH0
Zn in 9CH3
Zn in 9CH1