Zinc »
PDB 3m1q-3m6r »
3m6o »
Zinc in PDB 3m6o: Crystal Structure of Arabidopsis Thaliana Peptide Deformylase 1B (ATPDF1B)
Enzymatic activity of Crystal Structure of Arabidopsis Thaliana Peptide Deformylase 1B (ATPDF1B)
All present enzymatic activity of Crystal Structure of Arabidopsis Thaliana Peptide Deformylase 1B (ATPDF1B):
3.5.1.88;
3.5.1.88;
Protein crystallography data
The structure of Crystal Structure of Arabidopsis Thaliana Peptide Deformylase 1B (ATPDF1B), PDB code: 3m6o
was solved by
S.Fieulaine,
T.Meinnel,
C.Giglione,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 27.97 / 2.00 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 55.970, 55.940, 148.550, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 21.7 / 28.4 |
Zinc Binding Sites:
Pages:
>>> Page 1 <<< Page 2, Binding sites: 11 - 11;Binding sites:
The binding sites of Zinc atom in the Crystal Structure of Arabidopsis Thaliana Peptide Deformylase 1B (ATPDF1B) (pdb code 3m6o). This binding sites where shown within 5.0 Angstroms radius around Zinc atom.In total 11 binding sites of Zinc where determined in the Crystal Structure of Arabidopsis Thaliana Peptide Deformylase 1B (ATPDF1B), PDB code: 3m6o:
Jump to Zinc binding site number: 1; 2; 3; 4; 5; 6; 7; 8; 9; 10;
Zinc binding site 1 out of 11 in 3m6o
Go back to
Zinc binding site 1 out
of 11 in the Crystal Structure of Arabidopsis Thaliana Peptide Deformylase 1B (ATPDF1B)
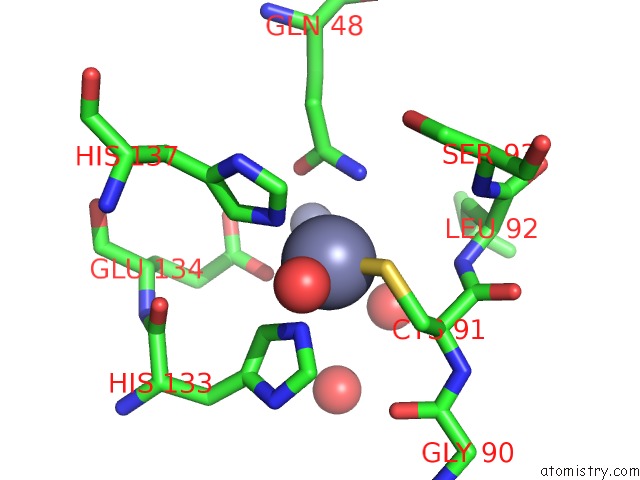
Mono view
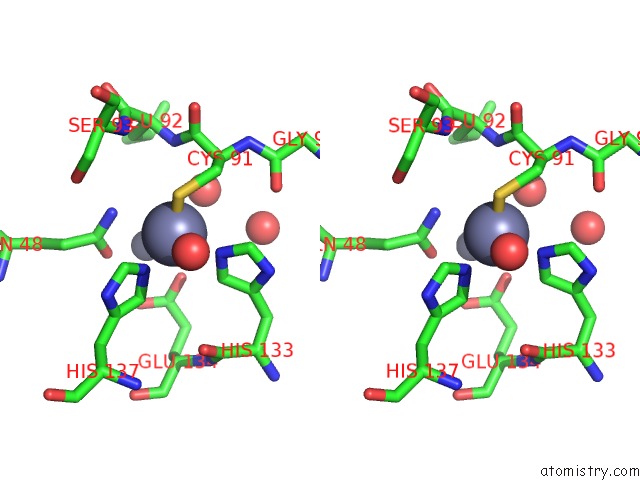
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Crystal Structure of Arabidopsis Thaliana Peptide Deformylase 1B (ATPDF1B) within 5.0Å range:
|
Zinc binding site 2 out of 11 in 3m6o
Go back to
Zinc binding site 2 out
of 11 in the Crystal Structure of Arabidopsis Thaliana Peptide Deformylase 1B (ATPDF1B)
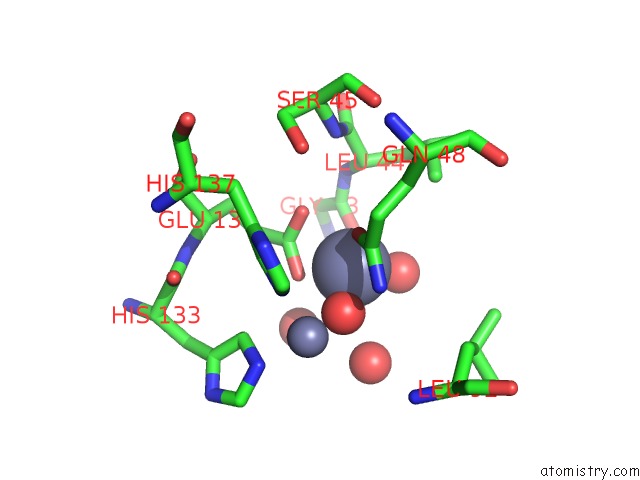
Mono view
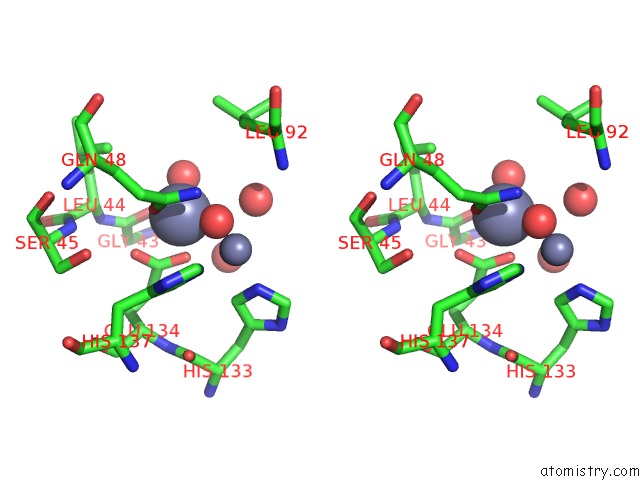
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of Crystal Structure of Arabidopsis Thaliana Peptide Deformylase 1B (ATPDF1B) within 5.0Å range:
|
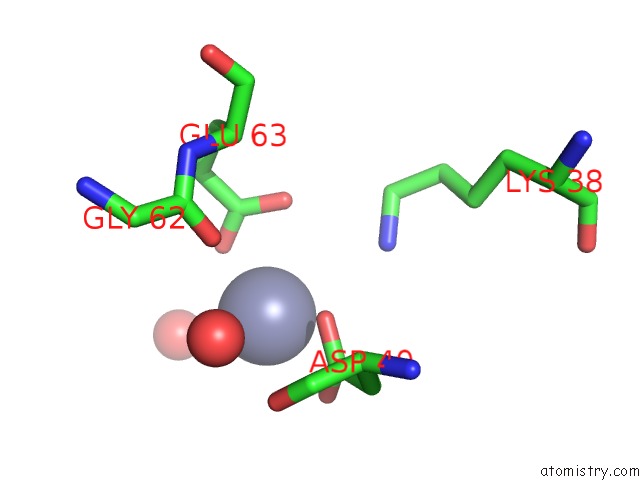
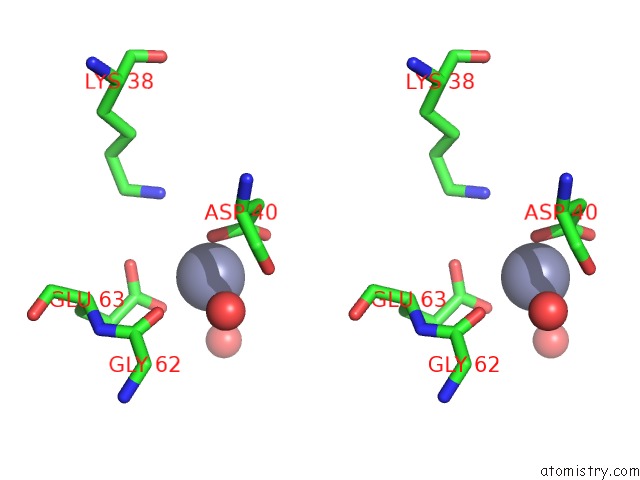
Zinc binding site 3 out of 11 in 3m6o
Go back to
Zinc binding site 3 out
of 11 in the Crystal Structure of Arabidopsis Thaliana Peptide Deformylase 1B (ATPDF1B)
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 3 of Crystal Structure of Arabidopsis Thaliana Peptide Deformylase 1B (ATPDF1B) within 5.0Å range:
|
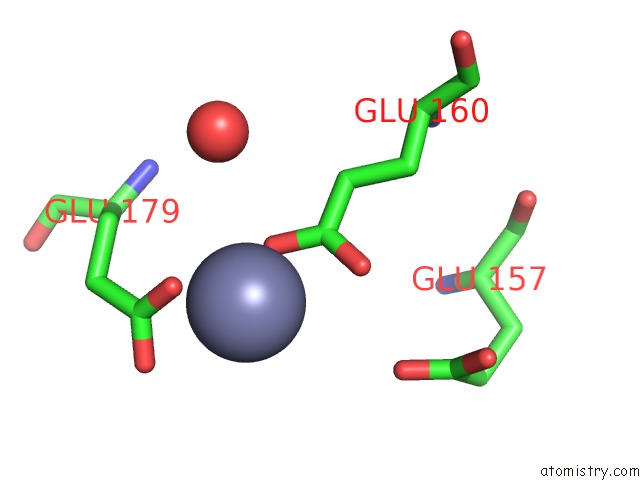
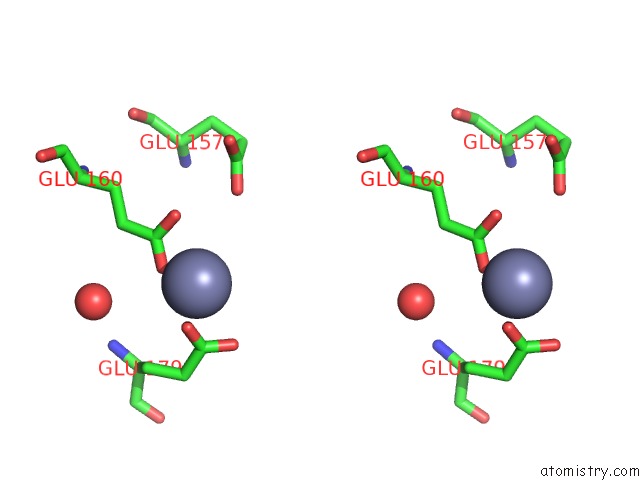
Zinc binding site 4 out of 11 in 3m6o
Go back to
Zinc binding site 4 out
of 11 in the Crystal Structure of Arabidopsis Thaliana Peptide Deformylase 1B (ATPDF1B)
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 4 of Crystal Structure of Arabidopsis Thaliana Peptide Deformylase 1B (ATPDF1B) within 5.0Å range:
|
Zinc binding site 5 out of 11 in 3m6o
Go back to
Zinc binding site 5 out
of 11 in the Crystal Structure of Arabidopsis Thaliana Peptide Deformylase 1B (ATPDF1B)
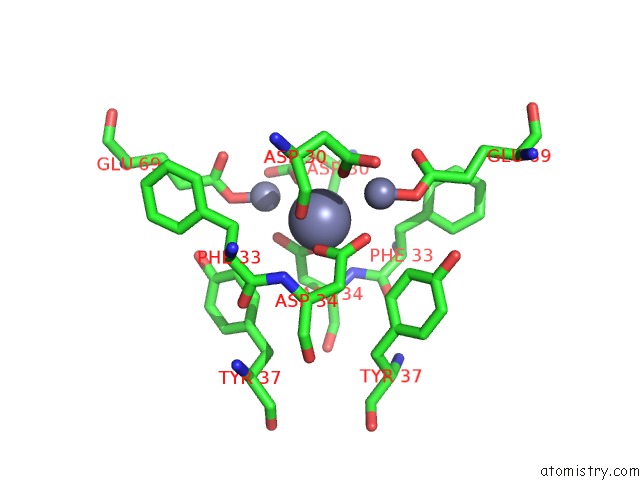
Mono view
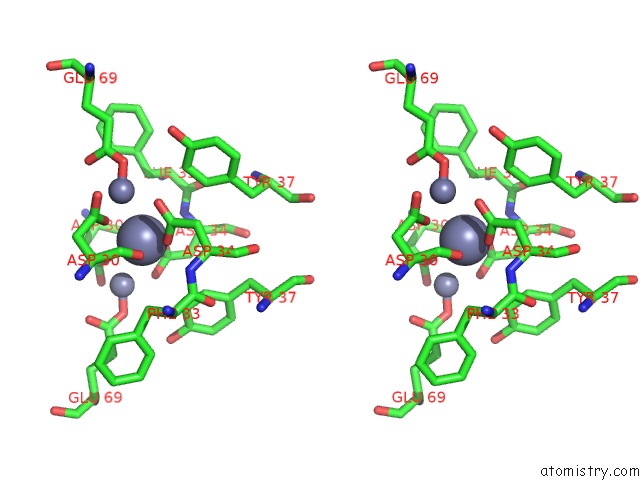
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 5 of Crystal Structure of Arabidopsis Thaliana Peptide Deformylase 1B (ATPDF1B) within 5.0Å range:
|
Zinc binding site 6 out of 11 in 3m6o
Go back to
Zinc binding site 6 out
of 11 in the Crystal Structure of Arabidopsis Thaliana Peptide Deformylase 1B (ATPDF1B)
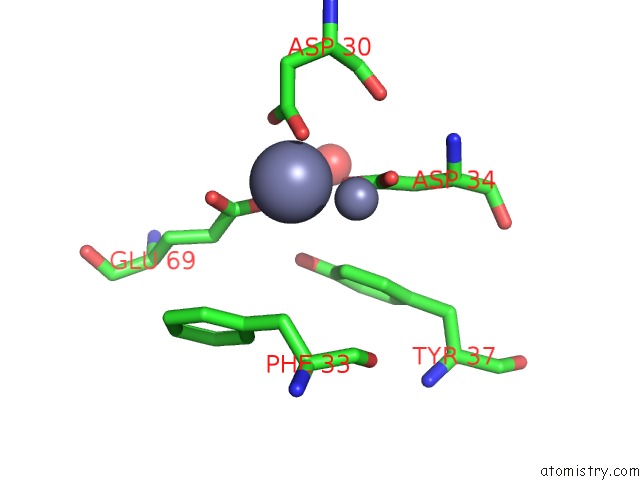
Mono view
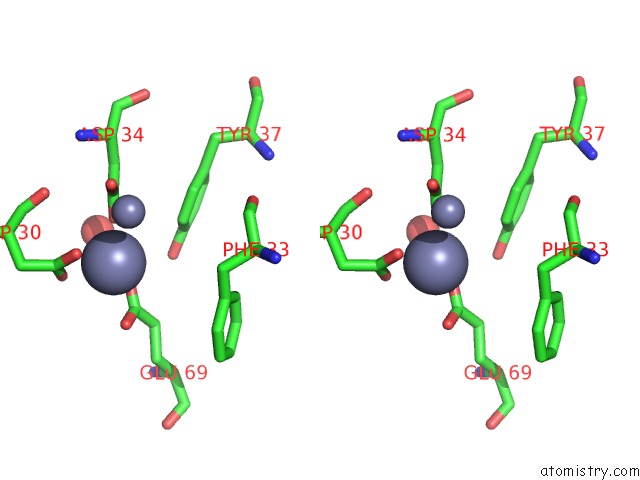
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 6 of Crystal Structure of Arabidopsis Thaliana Peptide Deformylase 1B (ATPDF1B) within 5.0Å range:
|
Zinc binding site 7 out of 11 in 3m6o
Go back to
Zinc binding site 7 out
of 11 in the Crystal Structure of Arabidopsis Thaliana Peptide Deformylase 1B (ATPDF1B)
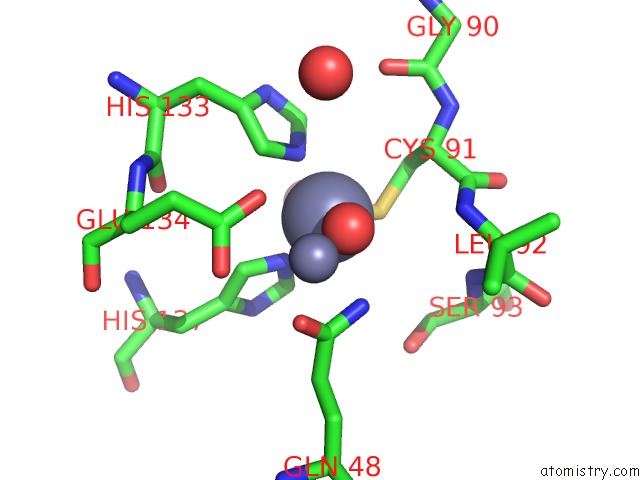
Mono view
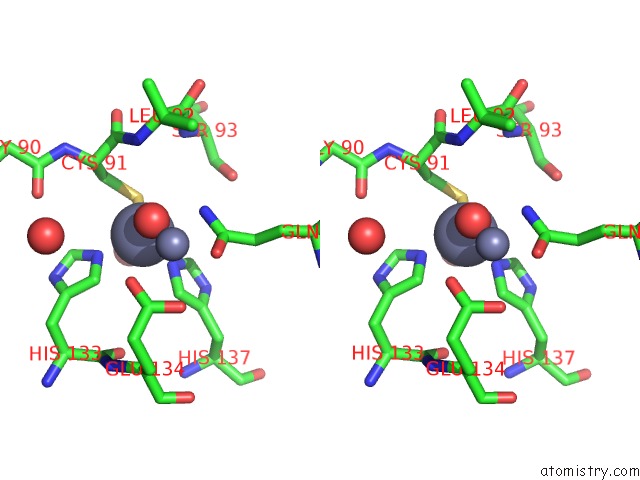
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 7 of Crystal Structure of Arabidopsis Thaliana Peptide Deformylase 1B (ATPDF1B) within 5.0Å range:
|
Zinc binding site 8 out of 11 in 3m6o
Go back to
Zinc binding site 8 out
of 11 in the Crystal Structure of Arabidopsis Thaliana Peptide Deformylase 1B (ATPDF1B)
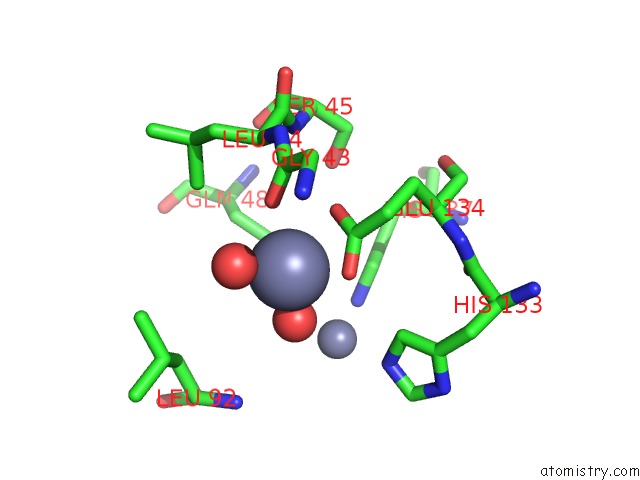
Mono view
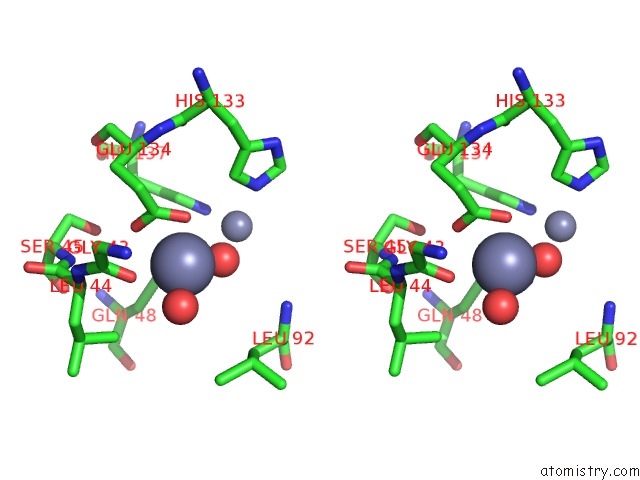
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 8 of Crystal Structure of Arabidopsis Thaliana Peptide Deformylase 1B (ATPDF1B) within 5.0Å range:
|
Zinc binding site 9 out of 11 in 3m6o
Go back to
Zinc binding site 9 out
of 11 in the Crystal Structure of Arabidopsis Thaliana Peptide Deformylase 1B (ATPDF1B)
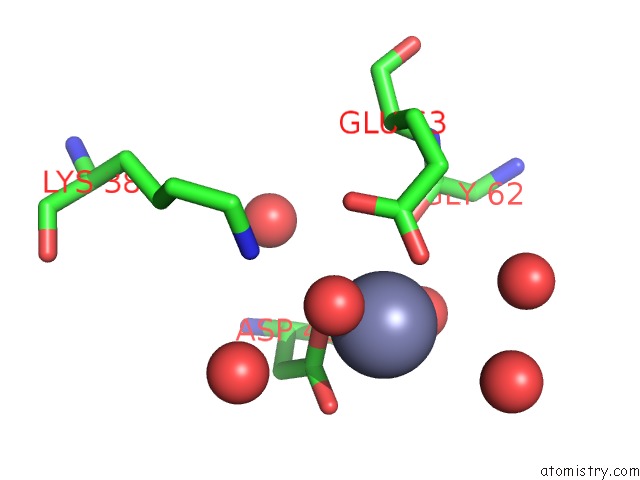
Mono view
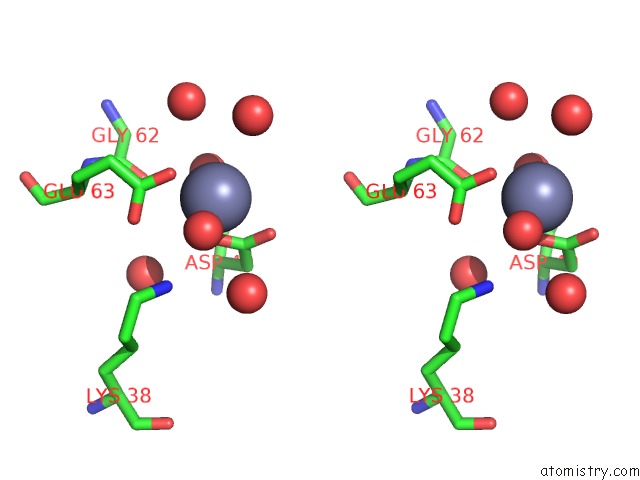
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 9 of Crystal Structure of Arabidopsis Thaliana Peptide Deformylase 1B (ATPDF1B) within 5.0Å range:
|
Zinc binding site 10 out of 11 in 3m6o
Go back to
Zinc binding site 10 out
of 11 in the Crystal Structure of Arabidopsis Thaliana Peptide Deformylase 1B (ATPDF1B)
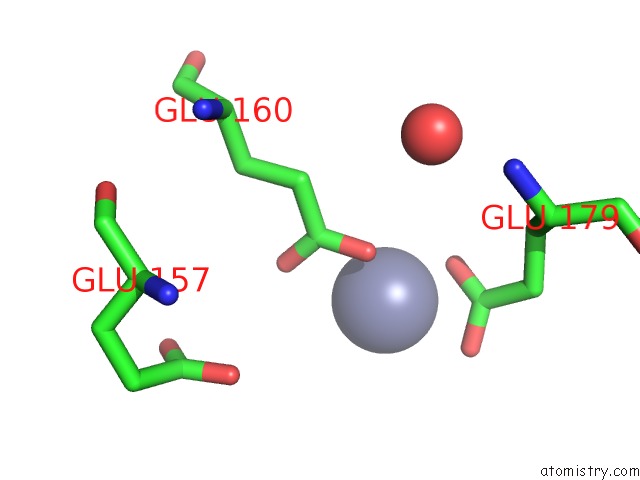
Mono view
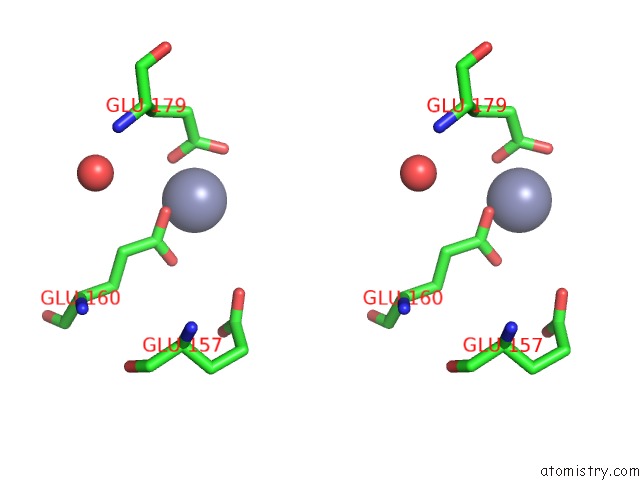
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 10 of Crystal Structure of Arabidopsis Thaliana Peptide Deformylase 1B (ATPDF1B) within 5.0Å range:
|
Reference:
S.Fieulaine,
A.Boularot,
I.Artaud,
M.Desmadril,
F.Dardel,
T.Meinnel,
C.Giglione.
Trapping Conformational States Along Ligand-Binding Dynamics of Peptide Deformylase: the Impact of Induced Fit on Enzyme Catalysis Plos Biol. V. 9 01066 2011.
ISSN: ISSN 1544-9173
PubMed: 21629676
DOI: 10.1371/JOURNAL.PBIO.1001066
Page generated: Wed Aug 20 11:40:40 2025
ISSN: ISSN 1544-9173
PubMed: 21629676
DOI: 10.1371/JOURNAL.PBIO.1001066
Last articles
Zn in 3TE7Zn in 3TDA
Zn in 3TBG
Zn in 3TDP
Zn in 3TC8
Zn in 3TAS
Zn in 3TAJ
Zn in 3T9U
Zn in 3T9X
Zn in 3TA7