Zinc »
PDB 3hk7-3hqz »
3hl5 »
Zinc in PDB 3hl5: Crystal Structure of Xiap BIR3 with CS3
Protein crystallography data
The structure of Crystal Structure of Xiap BIR3 with CS3, PDB code: 3hl5
was solved by
S.G.Hymowitz,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 49.15 / 1.80 |
| Space group | C 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 103.913, 39.940, 49.929, 90.00, 100.00, 90.00 |
| R / Rfree (%) | 17.6 / 20.9 |
Zinc Binding Sites:
The binding sites of Zinc atom in the Crystal Structure of Xiap BIR3 with CS3
(pdb code 3hl5). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 2 binding sites of Zinc where determined in the Crystal Structure of Xiap BIR3 with CS3, PDB code: 3hl5:
Jump to Zinc binding site number: 1; 2;
In total 2 binding sites of Zinc where determined in the Crystal Structure of Xiap BIR3 with CS3, PDB code: 3hl5:
Jump to Zinc binding site number: 1; 2;
Zinc binding site 1 out of 2 in 3hl5
Go back to
Zinc binding site 1 out
of 2 in the Crystal Structure of Xiap BIR3 with CS3
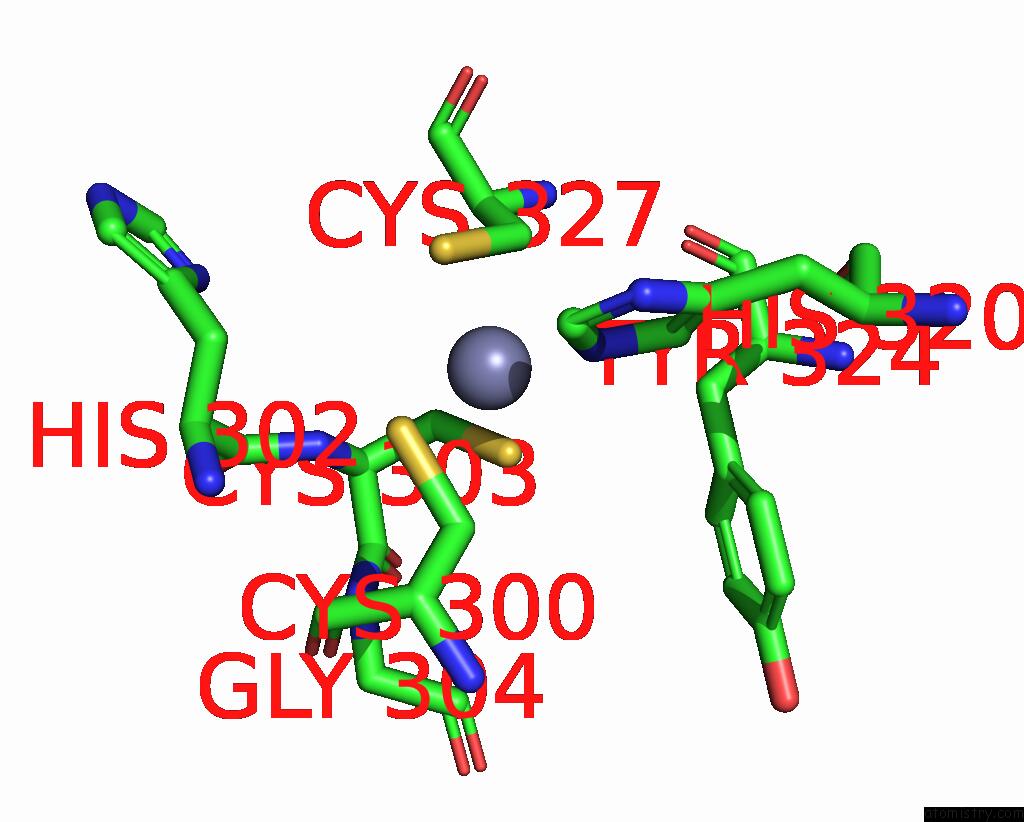
Mono view
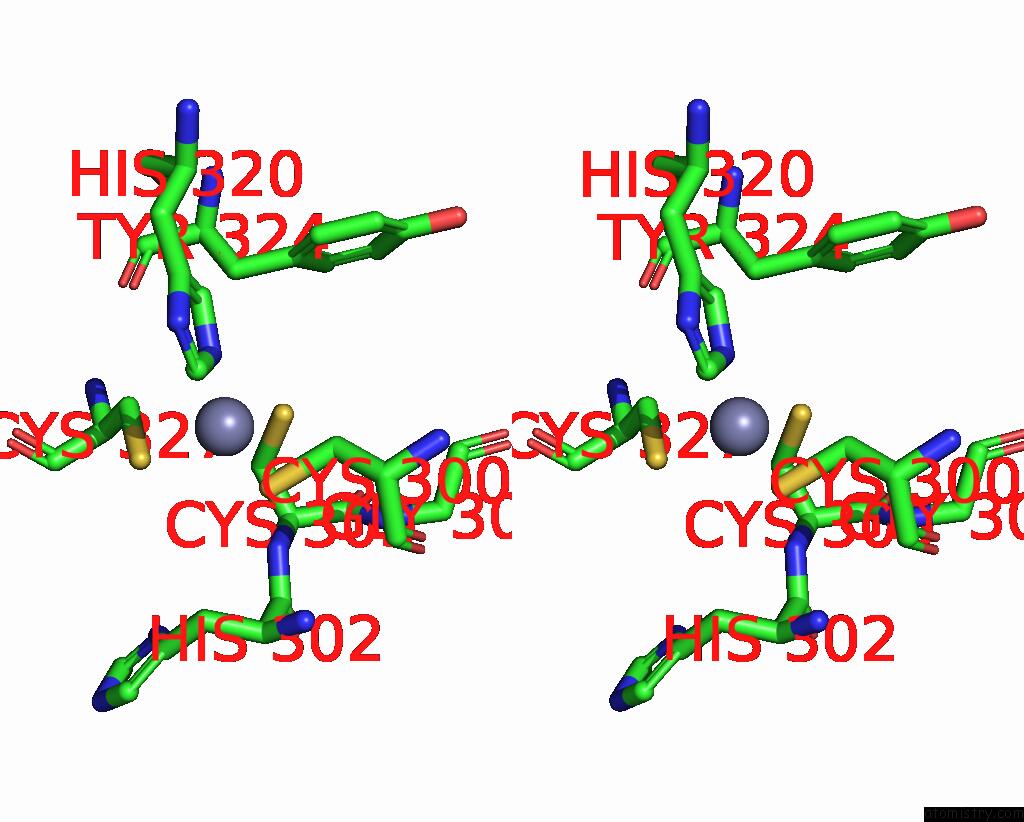
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Crystal Structure of Xiap BIR3 with CS3 within 5.0Å range:
|
Zinc binding site 2 out of 2 in 3hl5
Go back to
Zinc binding site 2 out
of 2 in the Crystal Structure of Xiap BIR3 with CS3
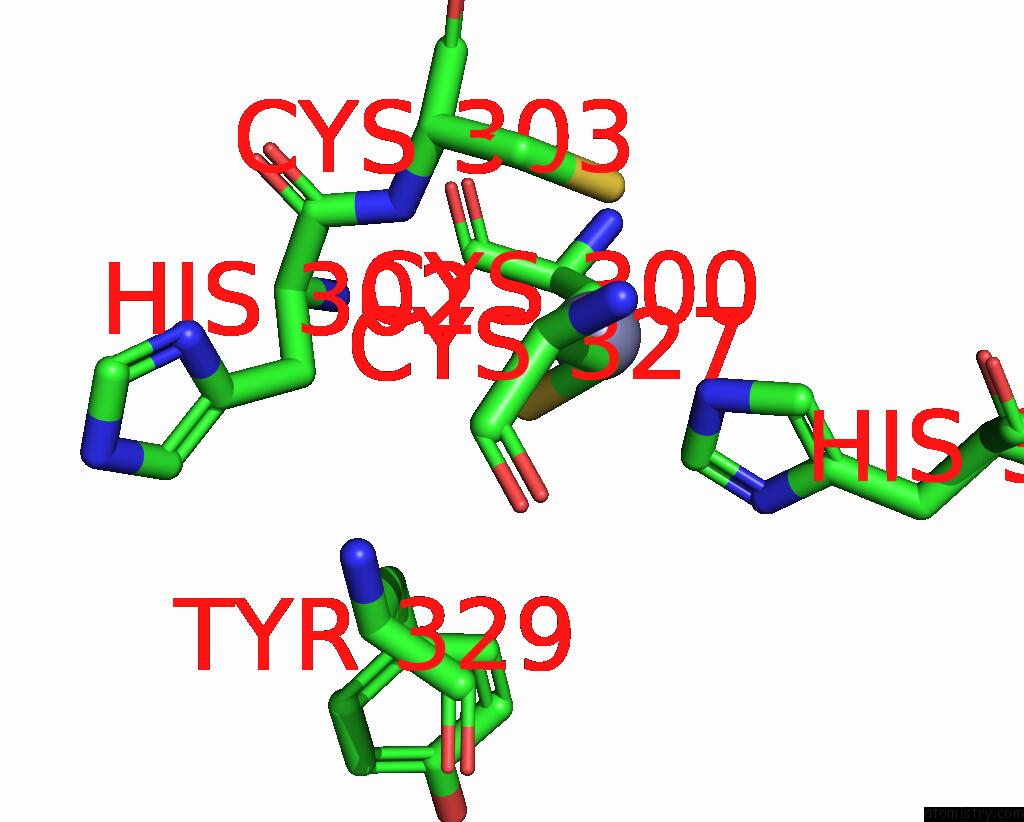
Mono view
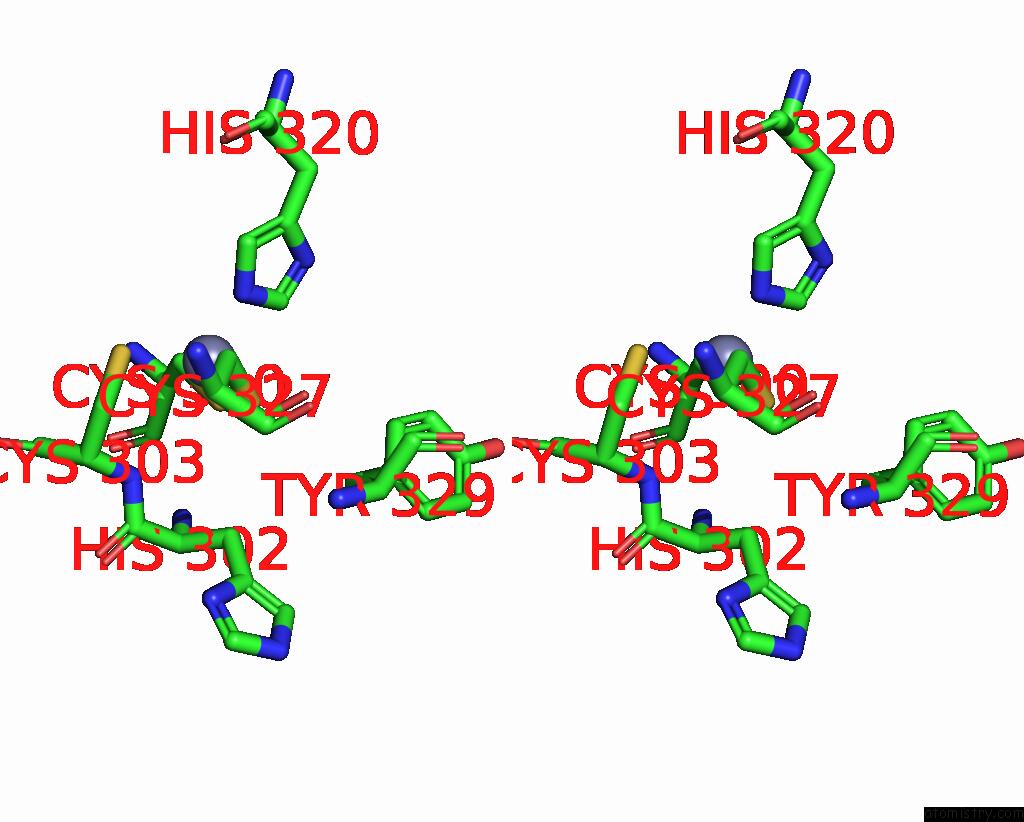
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of Crystal Structure of Xiap BIR3 with CS3 within 5.0Å range:
|
Reference:
C.Ndubaku,
E.Varfolomeev,
L.Wang,
K.Zobel,
K.Lau,
L.O.Elliott,
B.Maurer,
A.V.Fedorova,
J.N.Dynek,
M.Koehler,
S.G.Hymowitz,
V.Tsui,
K.Deshayes,
W.J.Fairbrother,
J.A.Flygare,
D.Vucic.
Antagonism of C-Iap and Xiap Proteins Is Required For Efficient Induction of Cell Death By Small-Molecule Iap Antagonists. Acs Chem.Biol. V. 4 557 2009.
ISSN: ISSN 1554-8929
PubMed: 19492850
DOI: 10.1021/CB900083M
Page generated: Wed Aug 20 10:07:16 2025
ISSN: ISSN 1554-8929
PubMed: 19492850
DOI: 10.1021/CB900083M
Last articles
Zn in 4DZHZn in 4DZ7
Zn in 4DZ9
Zn in 4DYO
Zn in 4DYG
Zn in 4DYK
Zn in 4DY1
Zn in 4DXH
Zn in 4DXX
Zn in 4DXU