Zinc »
PDB 3guc-3h69 »
3h0l »
Zinc in PDB 3h0l: Structure of Trna-Dependent Amidotransferase Gatcab From Aquifex Aeolicus
Protein crystallography data
The structure of Structure of Trna-Dependent Amidotransferase Gatcab From Aquifex Aeolicus, PDB code: 3h0l
was solved by
J.Wu,
W.Bu,
K.Sheppard,
M.Kitabatake,
D.Soll,
J.L.Smith,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 40.50 / 2.30 |
| Space group | P 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 127.482, 131.012, 154.668, 90.02, 90.00, 89.91 |
| R / Rfree (%) | 24 / 27.3 |
Other elements in 3h0l:
The structure of Structure of Trna-Dependent Amidotransferase Gatcab From Aquifex Aeolicus also contains other interesting chemical elements:
| Magnesium | (Mg) | 8 atoms |
Zinc Binding Sites:
The binding sites of Zinc atom in the Structure of Trna-Dependent Amidotransferase Gatcab From Aquifex Aeolicus
(pdb code 3h0l). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 8 binding sites of Zinc where determined in the Structure of Trna-Dependent Amidotransferase Gatcab From Aquifex Aeolicus, PDB code: 3h0l:
Jump to Zinc binding site number: 1; 2; 3; 4; 5; 6; 7; 8;
In total 8 binding sites of Zinc where determined in the Structure of Trna-Dependent Amidotransferase Gatcab From Aquifex Aeolicus, PDB code: 3h0l:
Jump to Zinc binding site number: 1; 2; 3; 4; 5; 6; 7; 8;
Zinc binding site 1 out of 8 in 3h0l
Go back to
Zinc binding site 1 out
of 8 in the Structure of Trna-Dependent Amidotransferase Gatcab From Aquifex Aeolicus
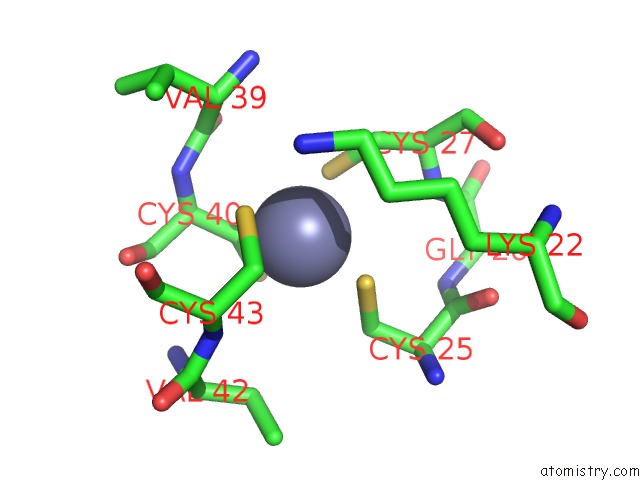
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Structure of Trna-Dependent Amidotransferase Gatcab From Aquifex Aeolicus within 5.0Å range:
|
Zinc binding site 2 out of 8 in 3h0l
Go back to
Zinc binding site 2 out
of 8 in the Structure of Trna-Dependent Amidotransferase Gatcab From Aquifex Aeolicus

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of Structure of Trna-Dependent Amidotransferase Gatcab From Aquifex Aeolicus within 5.0Å range:
|
Zinc binding site 3 out of 8 in 3h0l
Go back to
Zinc binding site 3 out
of 8 in the Structure of Trna-Dependent Amidotransferase Gatcab From Aquifex Aeolicus

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 3 of Structure of Trna-Dependent Amidotransferase Gatcab From Aquifex Aeolicus within 5.0Å range:
|
Zinc binding site 4 out of 8 in 3h0l
Go back to
Zinc binding site 4 out
of 8 in the Structure of Trna-Dependent Amidotransferase Gatcab From Aquifex Aeolicus
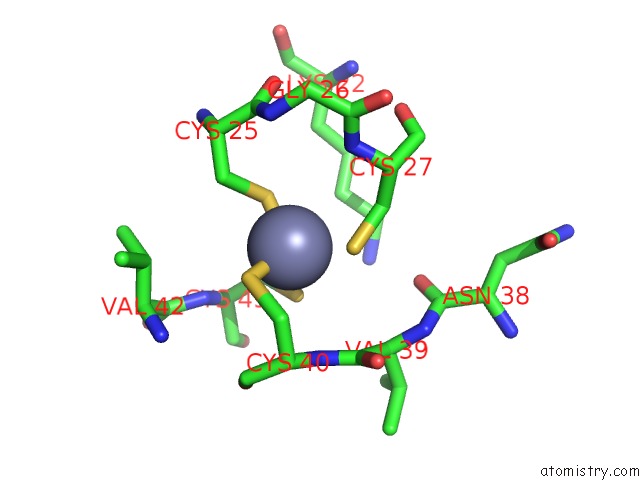
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 4 of Structure of Trna-Dependent Amidotransferase Gatcab From Aquifex Aeolicus within 5.0Å range:
|
Zinc binding site 5 out of 8 in 3h0l
Go back to
Zinc binding site 5 out
of 8 in the Structure of Trna-Dependent Amidotransferase Gatcab From Aquifex Aeolicus
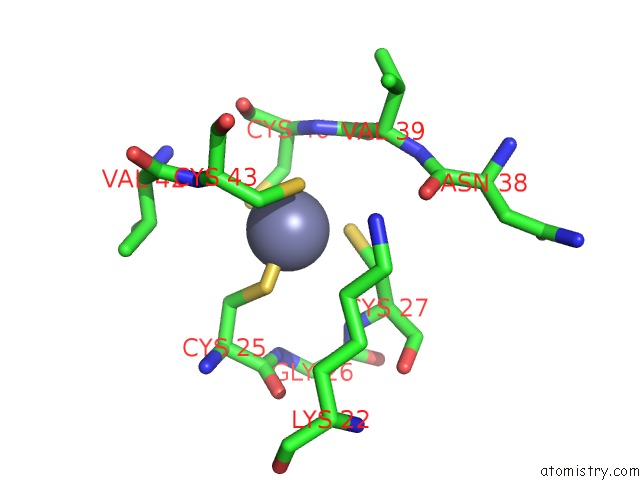
Mono view
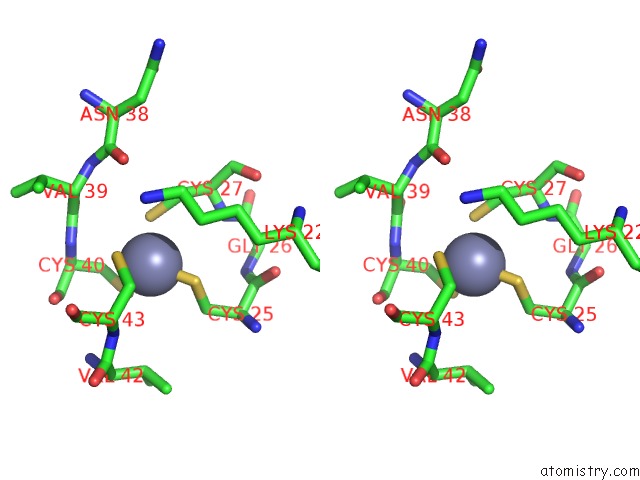
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 5 of Structure of Trna-Dependent Amidotransferase Gatcab From Aquifex Aeolicus within 5.0Å range:
|
Zinc binding site 6 out of 8 in 3h0l
Go back to
Zinc binding site 6 out
of 8 in the Structure of Trna-Dependent Amidotransferase Gatcab From Aquifex Aeolicus

Mono view
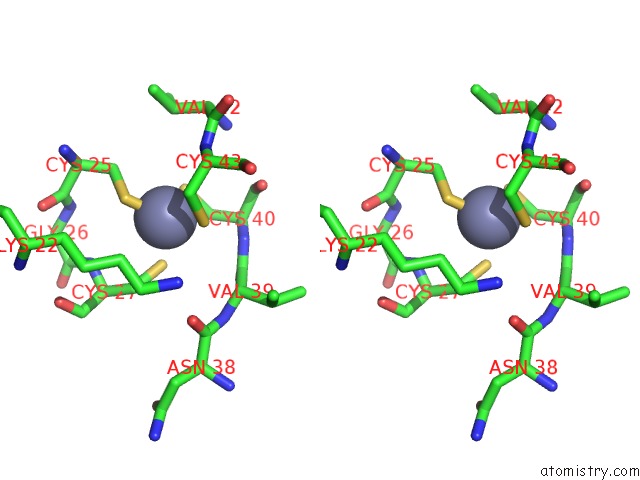
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 6 of Structure of Trna-Dependent Amidotransferase Gatcab From Aquifex Aeolicus within 5.0Å range:
|
Zinc binding site 7 out of 8 in 3h0l
Go back to
Zinc binding site 7 out
of 8 in the Structure of Trna-Dependent Amidotransferase Gatcab From Aquifex Aeolicus

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 7 of Structure of Trna-Dependent Amidotransferase Gatcab From Aquifex Aeolicus within 5.0Å range:
|
Zinc binding site 8 out of 8 in 3h0l
Go back to
Zinc binding site 8 out
of 8 in the Structure of Trna-Dependent Amidotransferase Gatcab From Aquifex Aeolicus

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 8 of Structure of Trna-Dependent Amidotransferase Gatcab From Aquifex Aeolicus within 5.0Å range:
|
Reference:
J.Wu,
W.Bu,
K.Sheppard,
M.Kitabatake,
S.T.Kwon,
D.Soll,
J.L.Smith.
Insights Into Trna-Dependent Amidotransferase Evolution and Catalysis From the Structure of the Aquifex Aeolicus Enzyme J.Mol.Biol. V. 391 703 2009.
ISSN: ISSN 0022-2836
PubMed: 19520089
DOI: 10.1016/J.JMB.2009.06.014
Page generated: Wed Aug 20 09:50:53 2025
ISSN: ISSN 0022-2836
PubMed: 19520089
DOI: 10.1016/J.JMB.2009.06.014
Last articles
Zn in 4BPDZn in 4BPB
Zn in 4BPA
Zn in 4BM9
Zn in 4BOL
Zn in 4BN4
Zn in 4BLD
Zn in 4BMB
Zn in 4BLL
Zn in 4BLB