Zinc »
PDB 3bof-3c52 »
3bup »
Zinc in PDB 3bup: Golgi Alpha-Mannosidase II D341N Acid-Base Catalyst Mutant with Bound Mannose
Enzymatic activity of Golgi Alpha-Mannosidase II D341N Acid-Base Catalyst Mutant with Bound Mannose
All present enzymatic activity of Golgi Alpha-Mannosidase II D341N Acid-Base Catalyst Mutant with Bound Mannose:
3.2.1.114;
3.2.1.114;
Protein crystallography data
The structure of Golgi Alpha-Mannosidase II D341N Acid-Base Catalyst Mutant with Bound Mannose, PDB code: 3bup
was solved by
D.A.Kuntz,
D.R.Rose,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 19.79 / 2.03 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 68.670, 109.486, 138.584, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 15.2 / 19.5 |
Zinc Binding Sites:
The binding sites of Zinc atom in the Golgi Alpha-Mannosidase II D341N Acid-Base Catalyst Mutant with Bound Mannose
(pdb code 3bup). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total only one binding site of Zinc was determined in the Golgi Alpha-Mannosidase II D341N Acid-Base Catalyst Mutant with Bound Mannose, PDB code: 3bup:
In total only one binding site of Zinc was determined in the Golgi Alpha-Mannosidase II D341N Acid-Base Catalyst Mutant with Bound Mannose, PDB code: 3bup:
Zinc binding site 1 out of 1 in 3bup
Go back to
Zinc binding site 1 out
of 1 in the Golgi Alpha-Mannosidase II D341N Acid-Base Catalyst Mutant with Bound Mannose
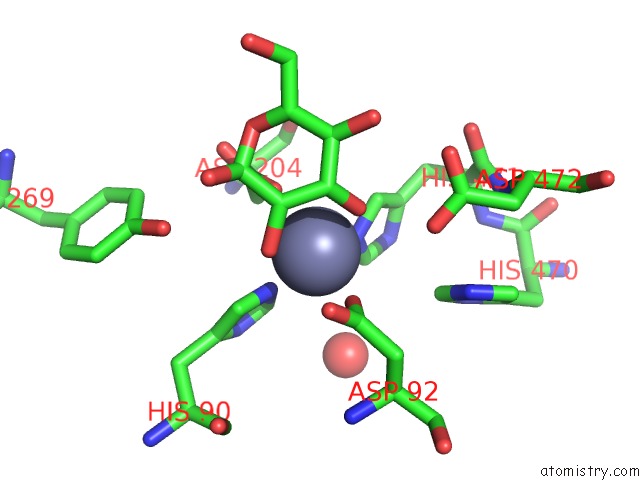
Mono view
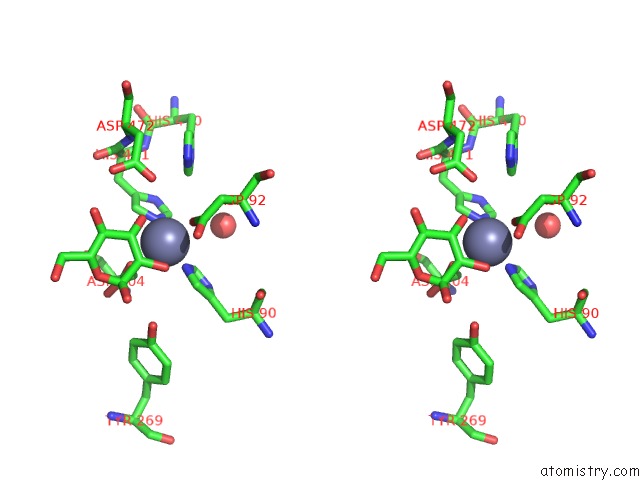
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Golgi Alpha-Mannosidase II D341N Acid-Base Catalyst Mutant with Bound Mannose within 5.0Å range:
|
Reference:
W.Zhong,
D.A.Kuntz,
B.Ember,
H.Singh,
K.W.Moremen,
D.R.Rose,
G.J.Boons.
Probing the Substrate Specificity of Golgi Alpha-Mannosidase II By Use of Synthetic Oligosaccharides and A Catalytic Nucleophile Mutant. J.Am.Chem.Soc. V. 130 8975 2008.
ISSN: ISSN 0002-7863
PubMed: 18558690
DOI: 10.1021/JA711248Y
Page generated: Wed Aug 20 08:04:44 2025
ISSN: ISSN 0002-7863
PubMed: 18558690
DOI: 10.1021/JA711248Y
Last articles
Zn in 3S73Zn in 3S72
Zn in 3S71
Zn in 3S5M
Zn in 3S5K
Zn in 3S3M
Zn in 3S3O
Zn in 3S3N
Zn in 3S32
Zn in 3S2I