Zinc »
PDB 2rgx-2rv5 »
2rhq »
Zinc in PDB 2rhq: Phers From Staphylococcus Haemolyticus- Rational Protein Engineering and Inhibitor Studies
Enzymatic activity of Phers From Staphylococcus Haemolyticus- Rational Protein Engineering and Inhibitor Studies
All present enzymatic activity of Phers From Staphylococcus Haemolyticus- Rational Protein Engineering and Inhibitor Studies:
6.1.1.20;
6.1.1.20;
Protein crystallography data
The structure of Phers From Staphylococcus Haemolyticus- Rational Protein Engineering and Inhibitor Studies, PDB code: 2rhq
was solved by
A.G.Evdokimov,
M.Mekel,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 35.60 / 2.20 |
| Space group | C 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 249.312, 87.602, 61.072, 90.00, 100.44, 90.00 |
| R / Rfree (%) | 19.6 / 26.9 |
Zinc Binding Sites:
The binding sites of Zinc atom in the Phers From Staphylococcus Haemolyticus- Rational Protein Engineering and Inhibitor Studies
(pdb code 2rhq). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total only one binding site of Zinc was determined in the Phers From Staphylococcus Haemolyticus- Rational Protein Engineering and Inhibitor Studies, PDB code: 2rhq:
In total only one binding site of Zinc was determined in the Phers From Staphylococcus Haemolyticus- Rational Protein Engineering and Inhibitor Studies, PDB code: 2rhq:
Zinc binding site 1 out of 1 in 2rhq
Go back to
Zinc binding site 1 out
of 1 in the Phers From Staphylococcus Haemolyticus- Rational Protein Engineering and Inhibitor Studies

Mono view
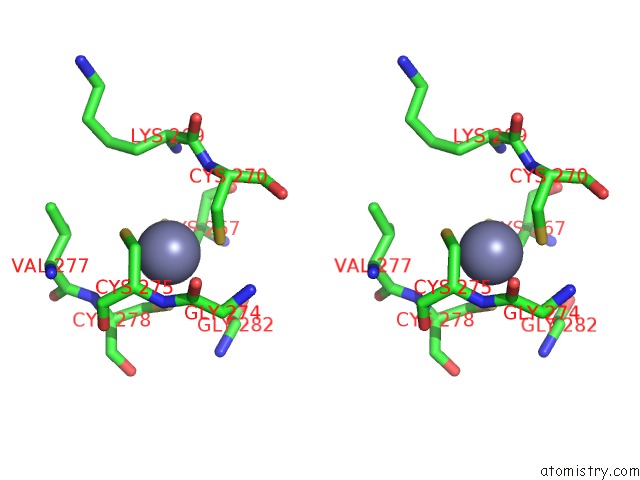
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Phers From Staphylococcus Haemolyticus- Rational Protein Engineering and Inhibitor Studies within 5.0Å range:
|
Reference:
A.G.Evdokimov,
M.Mekel,
K.Hutchings,
L.Narasimhan,
T.Holler,
T.Mcgrath,
B.Beattie,
E.Fauman,
C.Yan,
H.Heaslet,
R.Walter,
B.Finzel,
J.Ohren,
P.Mcconnell,
T.Braden,
F.Sun,
C.Spessard,
C.Banotai,
L.Al-Kassim,
W.Ma,
P.Wengender,
D.Kole,
N.Garceau,
P.Toogood,
J.Liu.
Rational Protein Engineering in Action: the First Crystal Structure of A Phenylalanine Trna Synthetase From Staphylococcus Haemolyticus. J.Struct.Biol. V. 162 152 2008.
ISSN: ISSN 1047-8477
PubMed: 18086534
DOI: 10.1016/J.JSB.2007.11.002
Page generated: Wed Aug 20 05:39:25 2025
ISSN: ISSN 1047-8477
PubMed: 18086534
DOI: 10.1016/J.JSB.2007.11.002
Last articles
Zn in 3KY9Zn in 3KWC
Zn in 3KRY
Zn in 3KWE
Zn in 3KWD
Zn in 3KWA
Zn in 3KVT
Zn in 3KVE
Zn in 3KV6
Zn in 3KV5