Zinc »
PDB 2jun-2kem »
2kdx »
Zinc in PDB 2kdx: Solution Structure of Hypa Protein
Zinc Binding Sites:
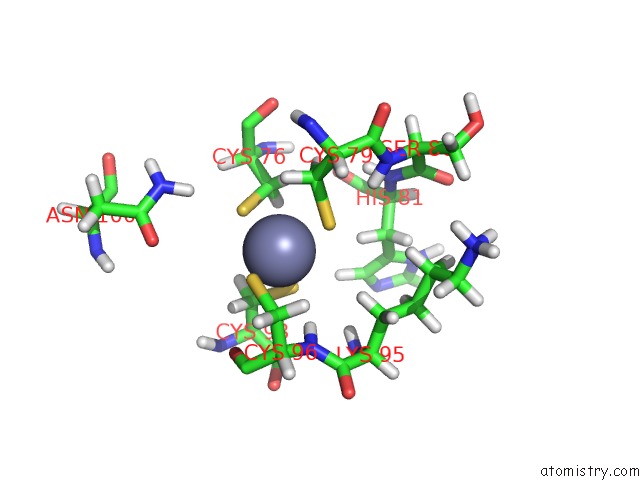
The binding sites of Zinc atom in the Solution Structure of Hypa Protein
(pdb code 2kdx). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total only one binding site of Zinc was determined in the Solution Structure of Hypa Protein, PDB code: 2kdx:
In total only one binding site of Zinc was determined in the Solution Structure of Hypa Protein, PDB code: 2kdx:
Zinc binding site 1 out of 1 in 2kdx
Go back to
Zinc binding site 1 out
of 1 in the Solution Structure of Hypa Protein
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Solution Structure of Hypa Protein within 5.0Å range:
|
Reference:
W.Xia,
H.Li,
K.-H.Sze,
H.Sun.
Structure of A Nickel Chaperone, Hypa, From Helicobacter Pylori Reveals Two Distinct Metal Binding Sites J.Am.Chem.Soc. V. 131 10031 2009.
ISSN: ISSN 0002-7863
PubMed: 19621959
DOI: 10.1021/JA900543Y
Page generated: Thu Oct 17 01:33:44 2024
ISSN: ISSN 0002-7863
PubMed: 19621959
DOI: 10.1021/JA900543Y
Last articles
Zn in 9J0NZn in 9J0O
Zn in 9J0P
Zn in 9FJX
Zn in 9EKB
Zn in 9C0F
Zn in 9CAH
Zn in 9CH0
Zn in 9CH3
Zn in 9CH1