Zinc »
PDB 2fac-2foq »
2fkm »
Zinc in PDB 2fkm: Pmm/Pgm S108D Mutant with Alpha-D-Glucose 1,6-Bisphosphate Bound
Enzymatic activity of Pmm/Pgm S108D Mutant with Alpha-D-Glucose 1,6-Bisphosphate Bound
All present enzymatic activity of Pmm/Pgm S108D Mutant with Alpha-D-Glucose 1,6-Bisphosphate Bound:
5.4.2.8;
5.4.2.8;
Protein crystallography data
The structure of Pmm/Pgm S108D Mutant with Alpha-D-Glucose 1,6-Bisphosphate Bound, PDB code: 2fkm
was solved by
C.A.Regni,
L.J.Beamer,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 42.83 / 1.90 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 70.222, 70.356, 84.392, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 20.3 / 25.1 |
Zinc Binding Sites:
The binding sites of Zinc atom in the Pmm/Pgm S108D Mutant with Alpha-D-Glucose 1,6-Bisphosphate Bound
(pdb code 2fkm). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total only one binding site of Zinc was determined in the Pmm/Pgm S108D Mutant with Alpha-D-Glucose 1,6-Bisphosphate Bound, PDB code: 2fkm:
In total only one binding site of Zinc was determined in the Pmm/Pgm S108D Mutant with Alpha-D-Glucose 1,6-Bisphosphate Bound, PDB code: 2fkm:
Zinc binding site 1 out of 1 in 2fkm
Go back to
Zinc binding site 1 out
of 1 in the Pmm/Pgm S108D Mutant with Alpha-D-Glucose 1,6-Bisphosphate Bound
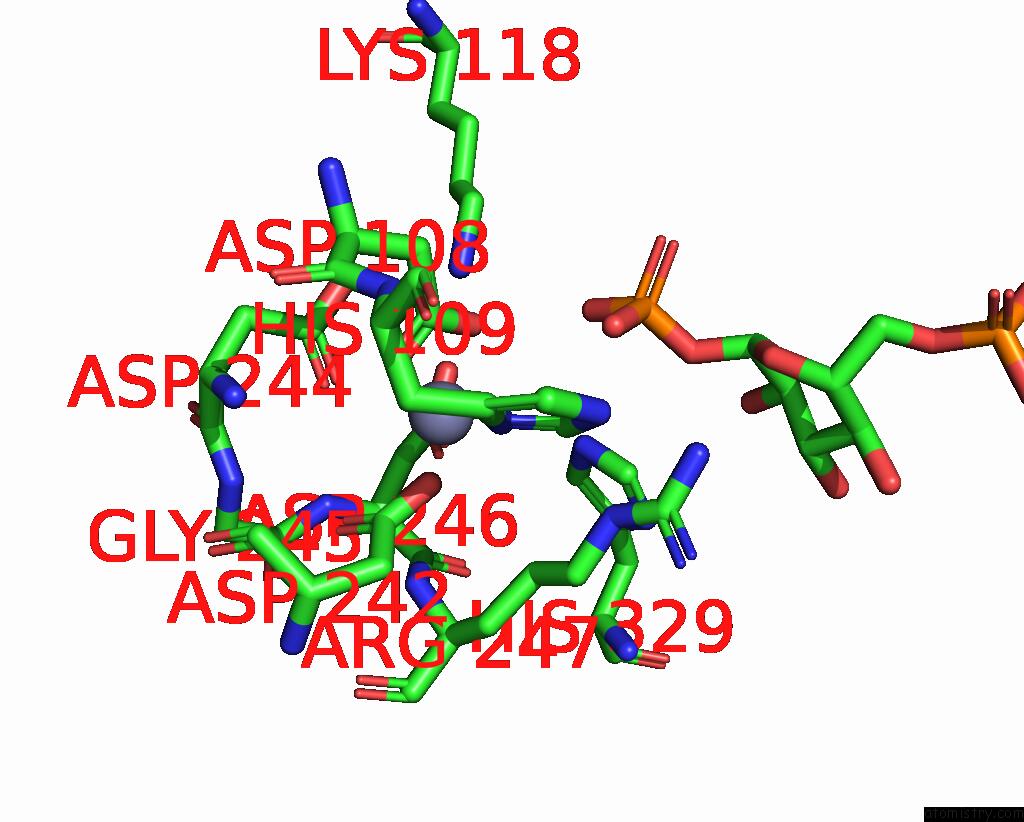
Mono view
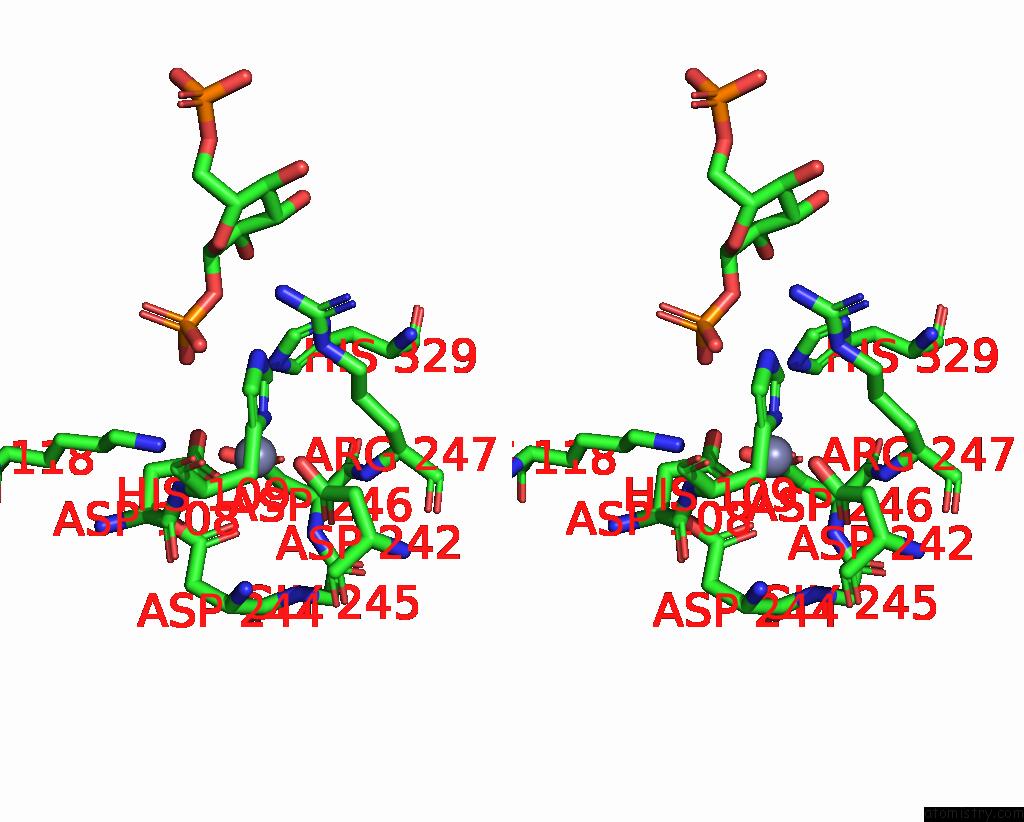
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Pmm/Pgm S108D Mutant with Alpha-D-Glucose 1,6-Bisphosphate Bound within 5.0Å range:
|
Reference:
C.Regni,
A.M.Schramm,
L.J.Beamer.
The Reaction of Phosphohexomutase From Pseudomonas Aeruginosa: Structural Insights Into A Simple Processive Enzyme. J.Biol.Chem. V. 281 15564 2006.
ISSN: ISSN 0021-9258
PubMed: 16595672
DOI: 10.1074/JBC.M600590200
Page generated: Wed Aug 20 02:44:46 2025
ISSN: ISSN 0021-9258
PubMed: 16595672
DOI: 10.1074/JBC.M600590200
Last articles
Zn in 2YWWZn in 2YU9
Zn in 2YVR
Zn in 2YW8
Zn in 2YUU
Zn in 2YV5
Zn in 2YUR
Zn in 2YUC
Zn in 2YU8
Zn in 2YU5