Zinc »
PDB 2ctt-2das »
2dar »
Zinc in PDB 2dar: Solution Structure of First Lim Domain of Enigma-Like Pdz and Lim Domains Protein
Zinc Binding Sites:
The binding sites of Zinc atom in the Solution Structure of First Lim Domain of Enigma-Like Pdz and Lim Domains Protein
(pdb code 2dar). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 2 binding sites of Zinc where determined in the Solution Structure of First Lim Domain of Enigma-Like Pdz and Lim Domains Protein, PDB code: 2dar:
Jump to Zinc binding site number: 1; 2;
In total 2 binding sites of Zinc where determined in the Solution Structure of First Lim Domain of Enigma-Like Pdz and Lim Domains Protein, PDB code: 2dar:
Jump to Zinc binding site number: 1; 2;
Zinc binding site 1 out of 2 in 2dar
Go back to
Zinc binding site 1 out
of 2 in the Solution Structure of First Lim Domain of Enigma-Like Pdz and Lim Domains Protein
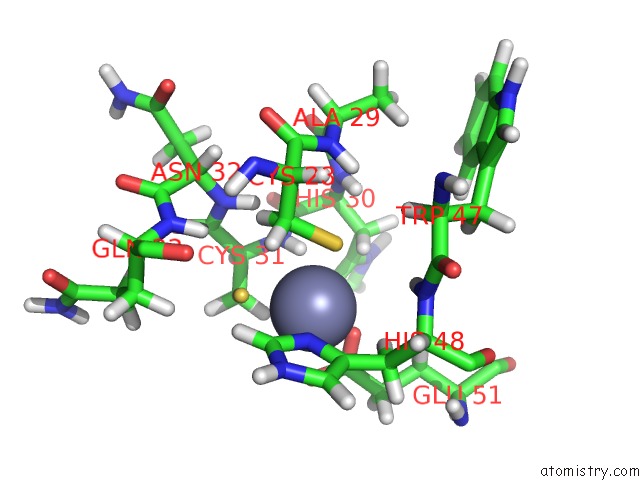
Mono view
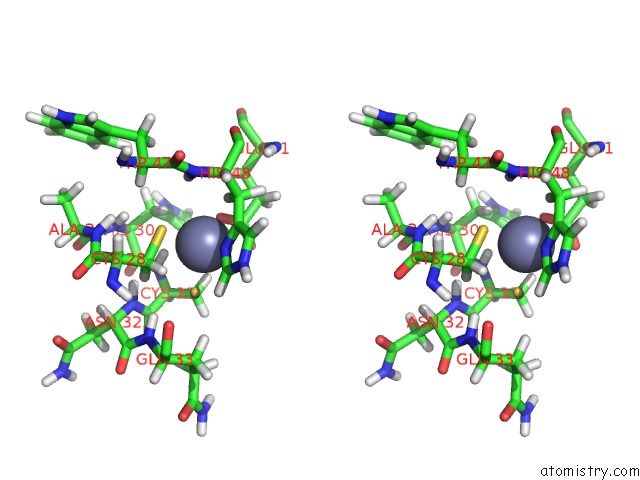
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Solution Structure of First Lim Domain of Enigma-Like Pdz and Lim Domains Protein within 5.0Å range:
|
Zinc binding site 2 out of 2 in 2dar
Go back to
Zinc binding site 2 out
of 2 in the Solution Structure of First Lim Domain of Enigma-Like Pdz and Lim Domains Protein
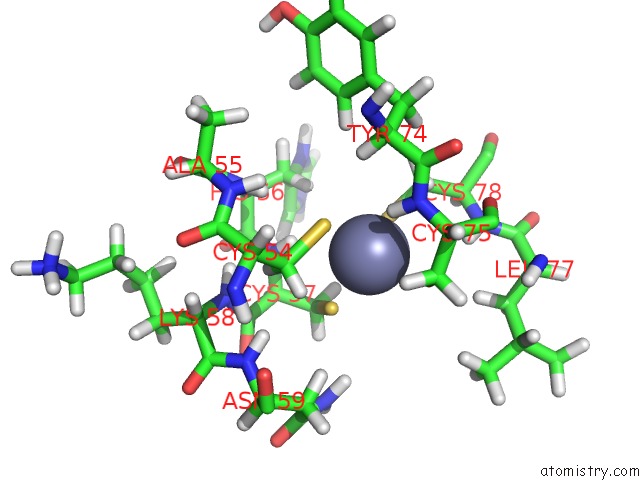
Mono view
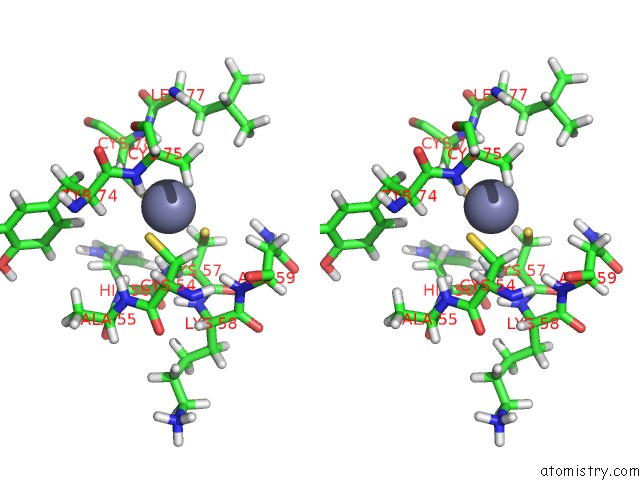
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of Solution Structure of First Lim Domain of Enigma-Like Pdz and Lim Domains Protein within 5.0Å range:
|
Reference:
T.N.Niraula,
Y.Muto,
S.Koshiba,
M.Inoue,
T.Kigawa,
S.Yokoyama.
Solution Structure of First Lim Domain of Enigma-Like Pdz and Lim Domains Protein To Be Published.
Page generated: Wed Aug 20 01:56:23 2025
Last articles
Zn in 2W44Zn in 2W2D
Zn in 2W3Q
Zn in 2W3Z
Zn in 2VXI
Zn in 2W0D
Zn in 2W22
Zn in 2W15
Zn in 2W14
Zn in 2W13