Zinc »
PDB 1zsc-2a2i »
1zu1 »
Zinc in PDB 1zu1: Solution Structure of the N-Terminal Zinc Fingers of the Xenopus Laevis Double Stranded Rna Binding Protein Zfa
Zinc Binding Sites:
The binding sites of Zinc atom in the Solution Structure of the N-Terminal Zinc Fingers of the Xenopus Laevis Double Stranded Rna Binding Protein Zfa
(pdb code 1zu1). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 2 binding sites of Zinc where determined in the Solution Structure of the N-Terminal Zinc Fingers of the Xenopus Laevis Double Stranded Rna Binding Protein Zfa, PDB code: 1zu1:
Jump to Zinc binding site number: 1; 2;
In total 2 binding sites of Zinc where determined in the Solution Structure of the N-Terminal Zinc Fingers of the Xenopus Laevis Double Stranded Rna Binding Protein Zfa, PDB code: 1zu1:
Jump to Zinc binding site number: 1; 2;
Zinc binding site 1 out of 2 in 1zu1
Go back to
Zinc binding site 1 out
of 2 in the Solution Structure of the N-Terminal Zinc Fingers of the Xenopus Laevis Double Stranded Rna Binding Protein Zfa
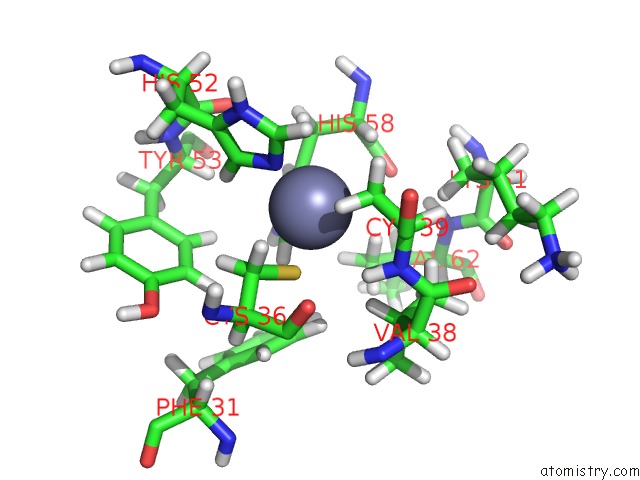
Mono view
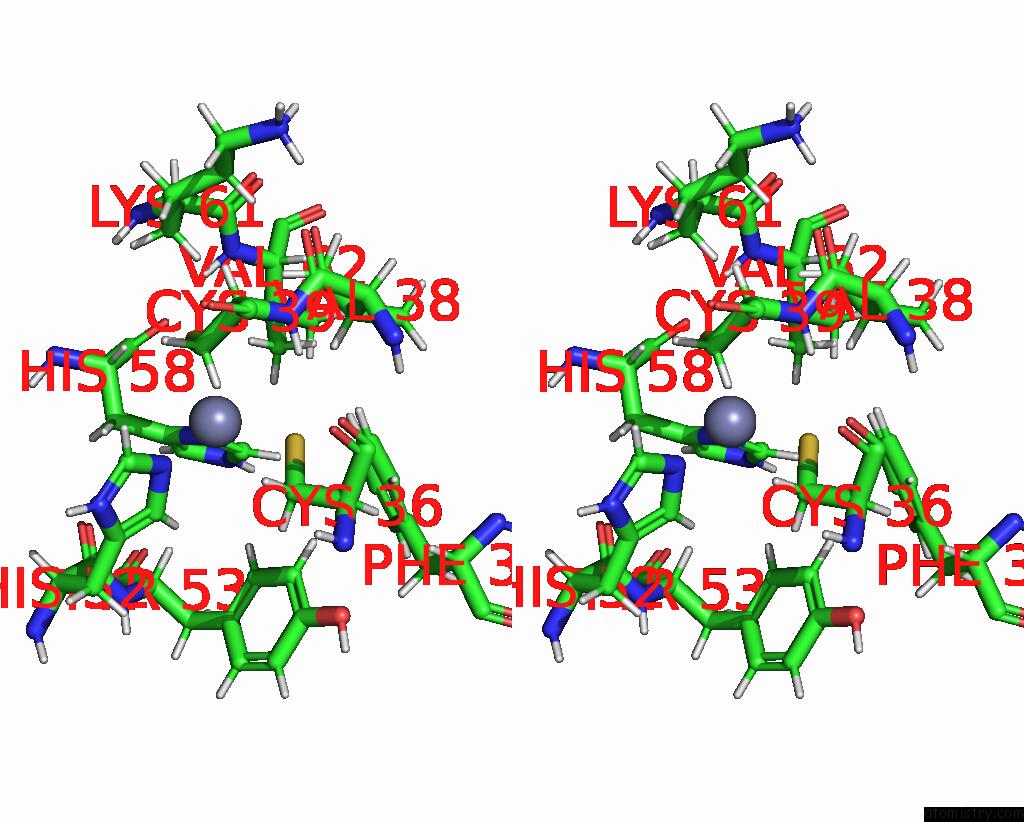
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Solution Structure of the N-Terminal Zinc Fingers of the Xenopus Laevis Double Stranded Rna Binding Protein Zfa within 5.0Å range:
|
Zinc binding site 2 out of 2 in 1zu1
Go back to
Zinc binding site 2 out
of 2 in the Solution Structure of the N-Terminal Zinc Fingers of the Xenopus Laevis Double Stranded Rna Binding Protein Zfa
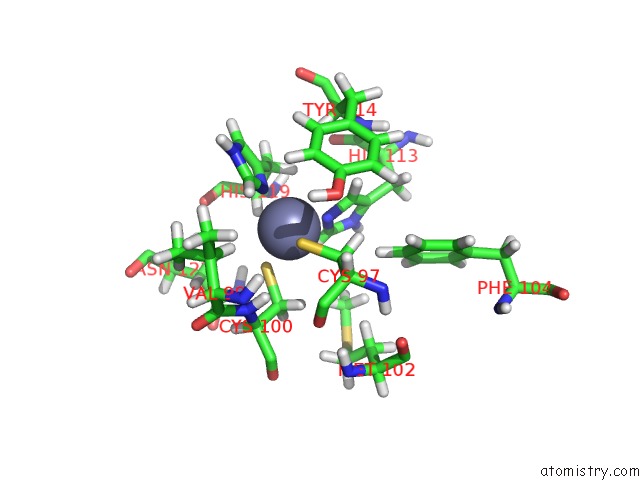
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of Solution Structure of the N-Terminal Zinc Fingers of the Xenopus Laevis Double Stranded Rna Binding Protein Zfa within 5.0Å range:
|
Reference:
H.M.Moller,
M.A.Martinez-Yamout,
H.J.Dyson,
P.E.Wright.
Solution Structure of the N-Terminal Zinc Fingers of the Xenopus Laevis Double-Stranded Rna-Binding Protein Zfa J.Mol.Biol. V. 351 718 2005.
ISSN: ISSN 0022-2836
PubMed: 16051273
DOI: 10.1016/J.JMB.2005.06.032
Page generated: Wed Oct 16 21:22:14 2024
ISSN: ISSN 0022-2836
PubMed: 16051273
DOI: 10.1016/J.JMB.2005.06.032
Last articles
Zn in 9J0NZn in 9J0O
Zn in 9J0P
Zn in 9FJX
Zn in 9EKB
Zn in 9C0F
Zn in 9CAH
Zn in 9CH0
Zn in 9CH3
Zn in 9CH1