Zinc »
PDB 1x8h-1xm8 »
1xjs »
Zinc in PDB 1xjs: Solution Structure of Iron-Sulfur Cluster Assembly Protein Iscu From Bacillus Subtilis, with Zinc Bound at the Active Site. Northeast Structural Genomics Consortium Target SR17
Zinc Binding Sites:
The binding sites of Zinc atom in the Solution Structure of Iron-Sulfur Cluster Assembly Protein Iscu From Bacillus Subtilis, with Zinc Bound at the Active Site. Northeast Structural Genomics Consortium Target SR17
(pdb code 1xjs). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total only one binding site of Zinc was determined in the Solution Structure of Iron-Sulfur Cluster Assembly Protein Iscu From Bacillus Subtilis, with Zinc Bound at the Active Site. Northeast Structural Genomics Consortium Target SR17, PDB code: 1xjs:
In total only one binding site of Zinc was determined in the Solution Structure of Iron-Sulfur Cluster Assembly Protein Iscu From Bacillus Subtilis, with Zinc Bound at the Active Site. Northeast Structural Genomics Consortium Target SR17, PDB code: 1xjs:
Zinc binding site 1 out of 1 in 1xjs
Go back to
Zinc binding site 1 out
of 1 in the Solution Structure of Iron-Sulfur Cluster Assembly Protein Iscu From Bacillus Subtilis, with Zinc Bound at the Active Site. Northeast Structural Genomics Consortium Target SR17
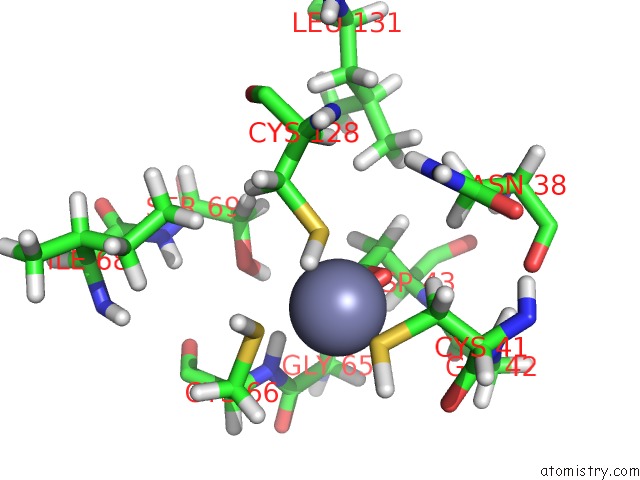
Mono view
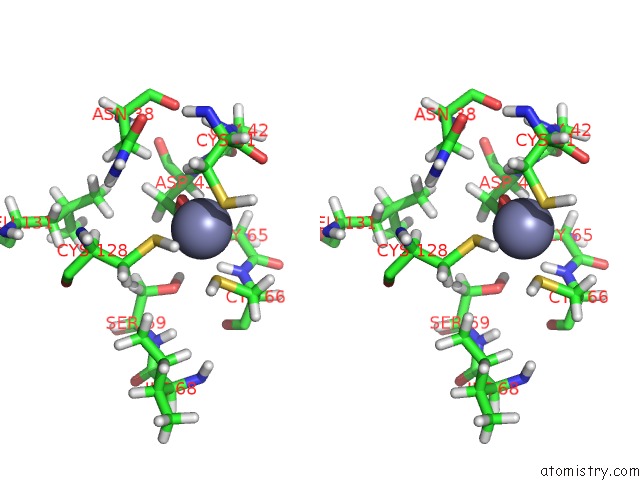
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Solution Structure of Iron-Sulfur Cluster Assembly Protein Iscu From Bacillus Subtilis, with Zinc Bound at the Active Site. Northeast Structural Genomics Consortium Target SR17 within 5.0Å range:
|
Reference:
G.J.Kornhaber,
G.V.T.Swapna,
T.A.Ramelot,
J.R.Cort,
M.A.Kennedy,
G.T.Montelione.
Solution Structure of Iron-Sulfur Cluster Assembly Protein Iscu From Bacillus Subtilis, with Zinc Bound at the Active Site. Northeast Structural Genomics Consortium Target SR17. To Be Published.
Page generated: Wed Aug 20 00:19:35 2025
Last articles
Zn in 2G9TZn in 2GDA
Zn in 2GC3
Zn in 2GD8
Zn in 2GC2
Zn in 2GC1
Zn in 2GC0
Zn in 2GBX
Zn in 2GBV
Zn in 2GBT