Zinc »
PDB 1oj7-1p1r »
1p0b »
Zinc in PDB 1p0b: Crystal Structure of Trna-Guanine Transglycosylase (Tgt) From Zymomonas Mobilis Complexed with Archaeosine Precursor, PREQ0
Enzymatic activity of Crystal Structure of Trna-Guanine Transglycosylase (Tgt) From Zymomonas Mobilis Complexed with Archaeosine Precursor, PREQ0
All present enzymatic activity of Crystal Structure of Trna-Guanine Transglycosylase (Tgt) From Zymomonas Mobilis Complexed with Archaeosine Precursor, PREQ0:
2.4.2.29;
2.4.2.29;
Protein crystallography data
The structure of Crystal Structure of Trna-Guanine Transglycosylase (Tgt) From Zymomonas Mobilis Complexed with Archaeosine Precursor, PREQ0, PDB code: 1p0b
was solved by
R.Brenk,
M.T.Stubbs,
A.Heine,
K.Reuter,
G.Klebe,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 20.00 / 1.70 |
| Space group | C 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 90.620, 65.540, 70.330, 90.00, 96.46, 90.00 |
| R / Rfree (%) | 17.6 / 20 |
Zinc Binding Sites:
The binding sites of Zinc atom in the Crystal Structure of Trna-Guanine Transglycosylase (Tgt) From Zymomonas Mobilis Complexed with Archaeosine Precursor, PREQ0
(pdb code 1p0b). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total only one binding site of Zinc was determined in the Crystal Structure of Trna-Guanine Transglycosylase (Tgt) From Zymomonas Mobilis Complexed with Archaeosine Precursor, PREQ0, PDB code: 1p0b:
In total only one binding site of Zinc was determined in the Crystal Structure of Trna-Guanine Transglycosylase (Tgt) From Zymomonas Mobilis Complexed with Archaeosine Precursor, PREQ0, PDB code: 1p0b:
Zinc binding site 1 out of 1 in 1p0b
Go back to
Zinc binding site 1 out
of 1 in the Crystal Structure of Trna-Guanine Transglycosylase (Tgt) From Zymomonas Mobilis Complexed with Archaeosine Precursor, PREQ0
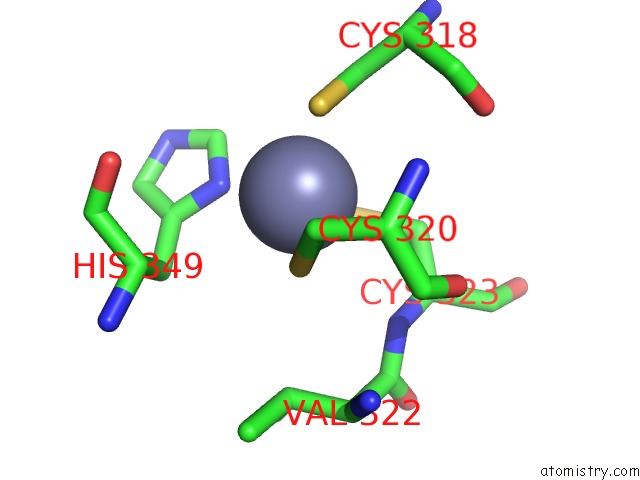
Mono view
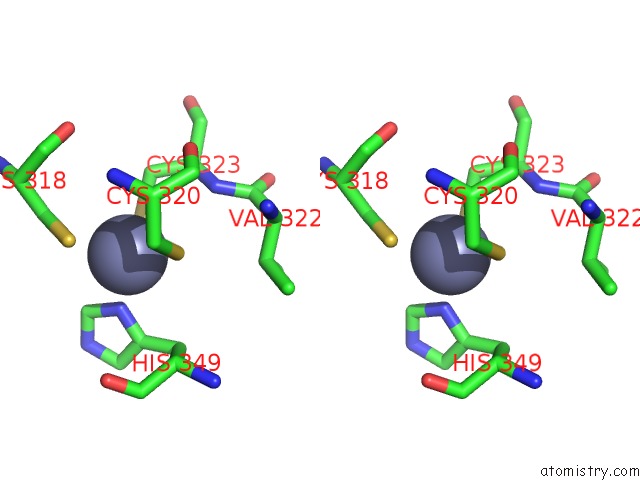
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Crystal Structure of Trna-Guanine Transglycosylase (Tgt) From Zymomonas Mobilis Complexed with Archaeosine Precursor, PREQ0 within 5.0Å range:
|
Reference:
R.Brenk,
M.T.Stubbs,
A.Heine,
K.Reuter,
G.Klebe.
Flexible Adaptations in the Structure of the Trna-Modifying Enzyme Trna-Guanine Transglycosylase and Their Implications For Substrate Selectivity, Reaction Mechanism and Structure-Based Drug Design Chembiochem V. 4 1066 2003.
ISSN: ISSN 1439-4227
PubMed: 14523925
DOI: 10.1002/CBIC.200300644
Page generated: Tue Aug 19 22:15:09 2025
ISSN: ISSN 1439-4227
PubMed: 14523925
DOI: 10.1002/CBIC.200300644
Last articles
Zn in 2A0SZn in 2A0B
Zn in 1ZZU
Zn in 258L
Zn in 2A03
Zn in 1ZZM
Zn in 1ZZS
Zn in 1ZZT
Zn in 1ZZR
Zn in 1ZZQ