Zinc »
PDB 9b12-9bz0 »
9b85 »
Zinc in PDB 9b85: Cryo-Em Structure of Human Dynactin Complex Bound to Chlamydia Effector DRE1
Zinc Binding Sites:
The binding sites of Zinc atom in the Cryo-Em Structure of Human Dynactin Complex Bound to Chlamydia Effector DRE1
(pdb code 9b85). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 3 binding sites of Zinc where determined in the Cryo-Em Structure of Human Dynactin Complex Bound to Chlamydia Effector DRE1, PDB code: 9b85:
Jump to Zinc binding site number: 1; 2; 3;
In total 3 binding sites of Zinc where determined in the Cryo-Em Structure of Human Dynactin Complex Bound to Chlamydia Effector DRE1, PDB code: 9b85:
Jump to Zinc binding site number: 1; 2; 3;
Zinc binding site 1 out of 3 in 9b85
Go back to
Zinc binding site 1 out
of 3 in the Cryo-Em Structure of Human Dynactin Complex Bound to Chlamydia Effector DRE1
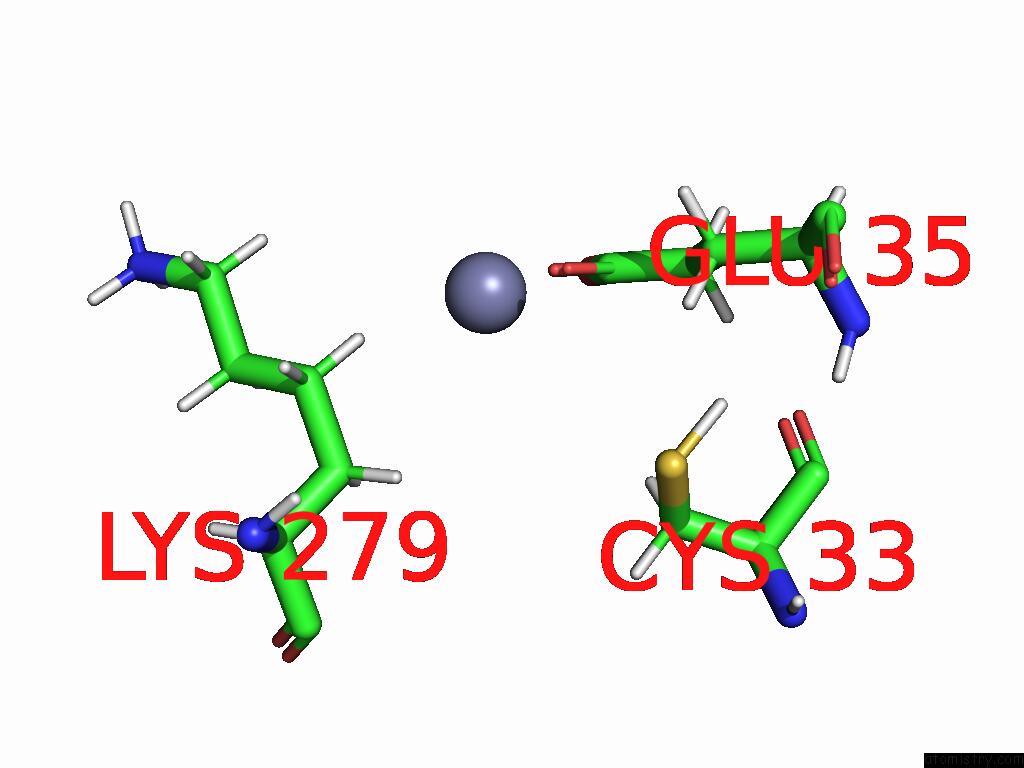
Mono view
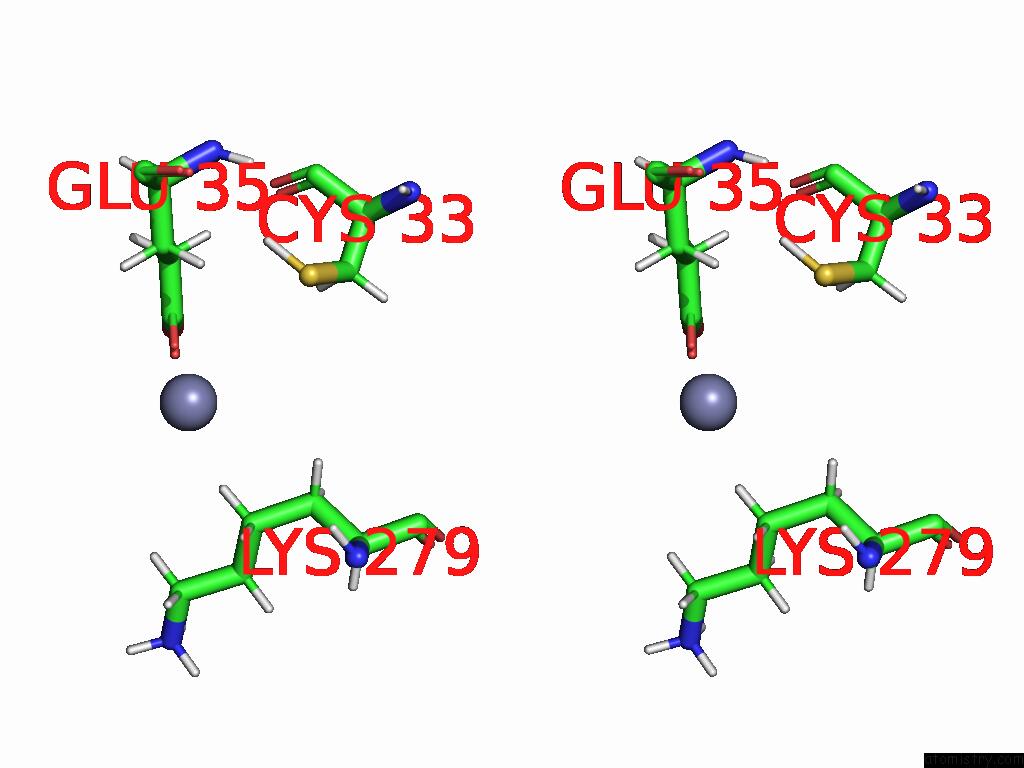
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Cryo-Em Structure of Human Dynactin Complex Bound to Chlamydia Effector DRE1 within 5.0Å range:
|
Zinc binding site 2 out of 3 in 9b85
Go back to
Zinc binding site 2 out
of 3 in the Cryo-Em Structure of Human Dynactin Complex Bound to Chlamydia Effector DRE1
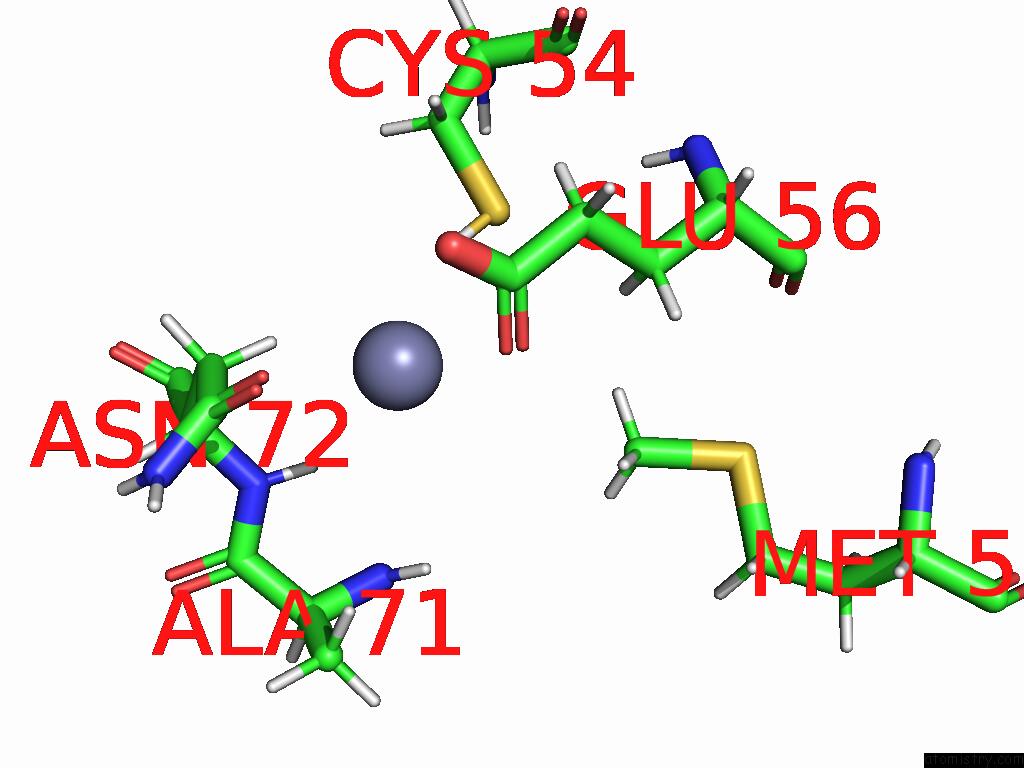
Mono view
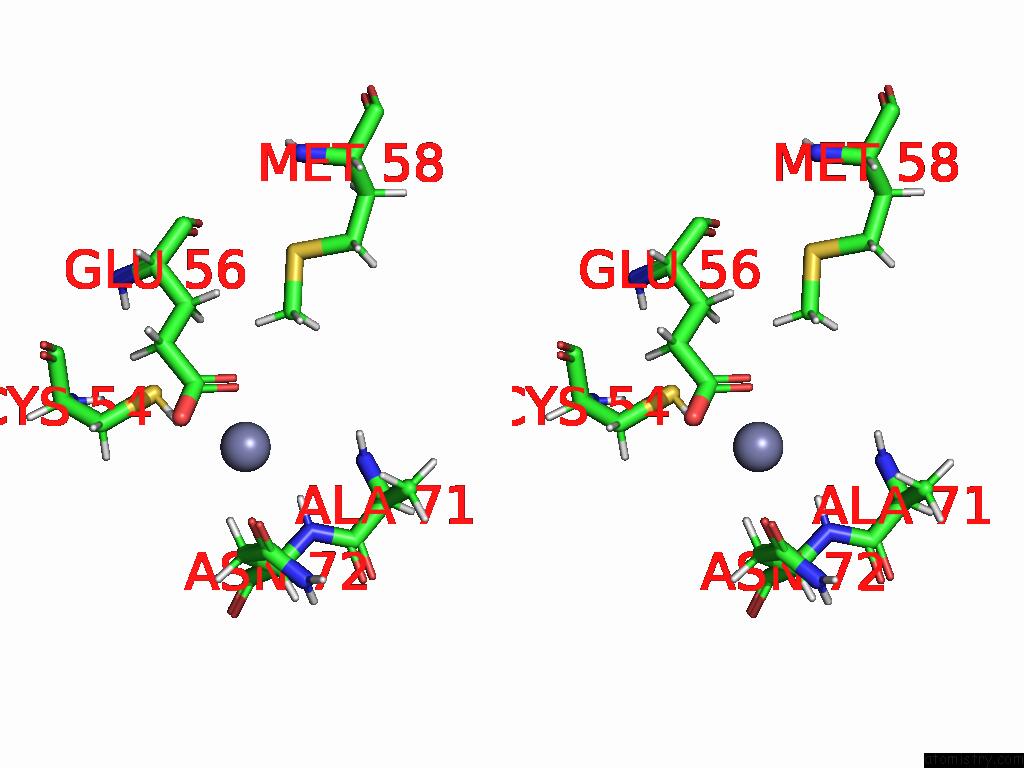
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of Cryo-Em Structure of Human Dynactin Complex Bound to Chlamydia Effector DRE1 within 5.0Å range:
|
Zinc binding site 3 out of 3 in 9b85
Go back to
Zinc binding site 3 out
of 3 in the Cryo-Em Structure of Human Dynactin Complex Bound to Chlamydia Effector DRE1
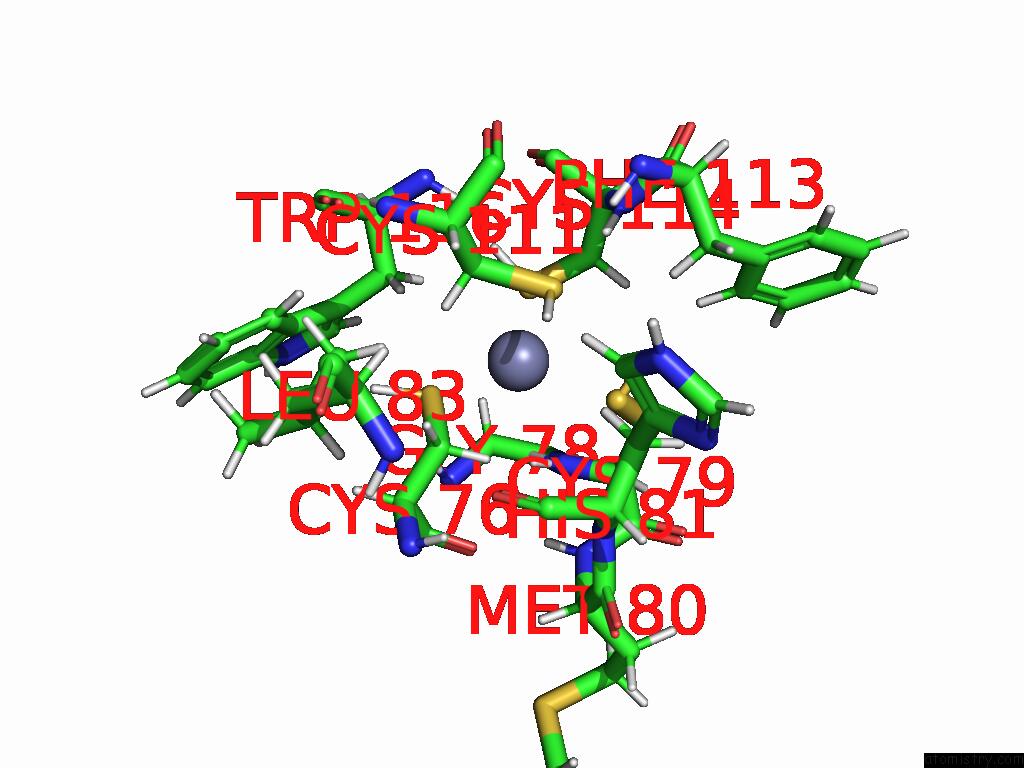
Mono view
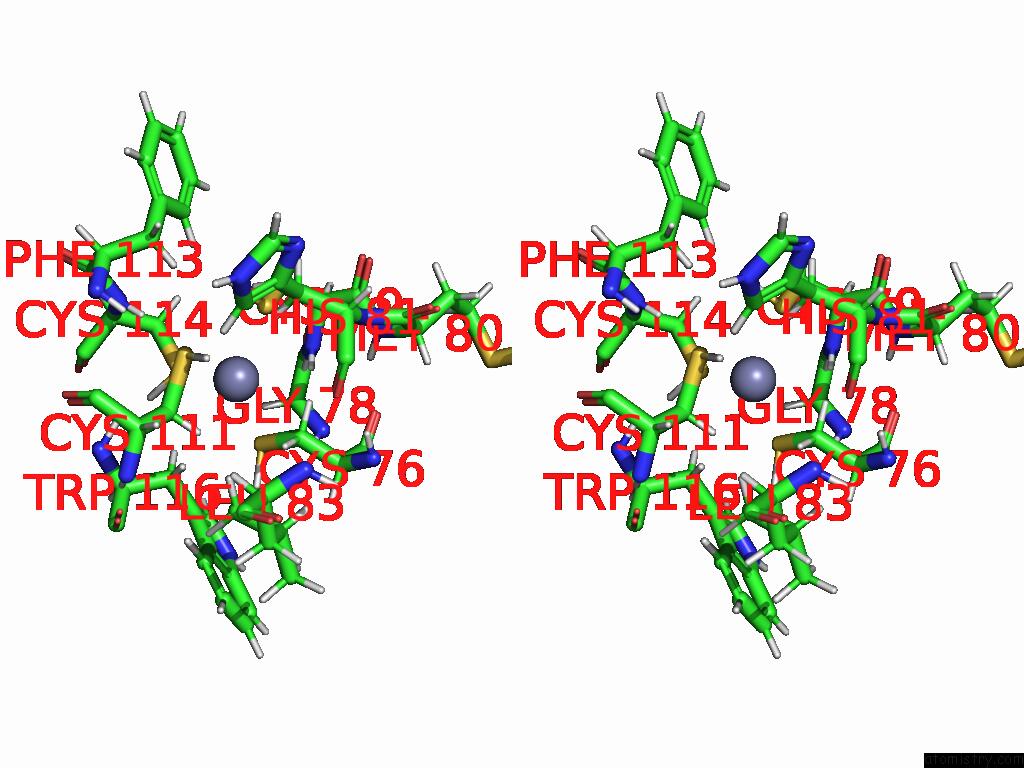
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 3 of Cryo-Em Structure of Human Dynactin Complex Bound to Chlamydia Effector DRE1 within 5.0Å range:
|
Reference:
J.Sherry,
K.I.Pawar,
L.Dolat,
E.Smith,
I.C.Chang,
K.Pha,
R.Kaake,
D.L.Swaney,
C.Herrera,
E.Mcmahon,
R.J.Bastidas,
J.R.Johnson,
R.H.Valdivia,
N.J.Krogan,
C.A.Elwell,
K.Verba,
J.N.Engel.
The Chlamydia Effector DRE1 Binds Dynactin to Reposition Host Organelles During Infection. Cell Rep V. 44 15509 2025.
ISSN: ESSN 2211-1247
PubMed: 40186871
DOI: 10.1016/J.CELREP.2025.115509
Page generated: Fri Aug 22 16:21:22 2025
ISSN: ESSN 2211-1247
PubMed: 40186871
DOI: 10.1016/J.CELREP.2025.115509
Last articles
Au in 9D33As in 9O9I
Al in 9GSG
Zr in 1XC1
Zr in 6Y7P
Zr in 6GNL
Zr in 6HYB
Zr in 4XYY
Zr in 5KHP
Zn in 9VXG