Zinc »
PDB 6sj4-6su5 »
6spn »
Zinc in PDB 6spn: Structure of the Escherichia Coli Methionyl-Trna Synthetase Complexed with Beta-Methionine
Enzymatic activity of Structure of the Escherichia Coli Methionyl-Trna Synthetase Complexed with Beta-Methionine
All present enzymatic activity of Structure of the Escherichia Coli Methionyl-Trna Synthetase Complexed with Beta-Methionine:
6.1.1.10;
6.1.1.10;
Protein crystallography data
The structure of Structure of the Escherichia Coli Methionyl-Trna Synthetase Complexed with Beta-Methionine, PDB code: 6spn
was solved by
G.Nigro,
E.Schmitt,
Y.Mechulam,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 41.69 / 1.45 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 78.225, 45.230, 86.252, 90.00, 107.36, 90.00 |
| R / Rfree (%) | 9.3 / 12.6 |
Zinc Binding Sites:
The binding sites of Zinc atom in the Structure of the Escherichia Coli Methionyl-Trna Synthetase Complexed with Beta-Methionine
(pdb code 6spn). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total only one binding site of Zinc was determined in the Structure of the Escherichia Coli Methionyl-Trna Synthetase Complexed with Beta-Methionine, PDB code: 6spn:
In total only one binding site of Zinc was determined in the Structure of the Escherichia Coli Methionyl-Trna Synthetase Complexed with Beta-Methionine, PDB code: 6spn:
Zinc binding site 1 out of 1 in 6spn
Go back to
Zinc binding site 1 out
of 1 in the Structure of the Escherichia Coli Methionyl-Trna Synthetase Complexed with Beta-Methionine
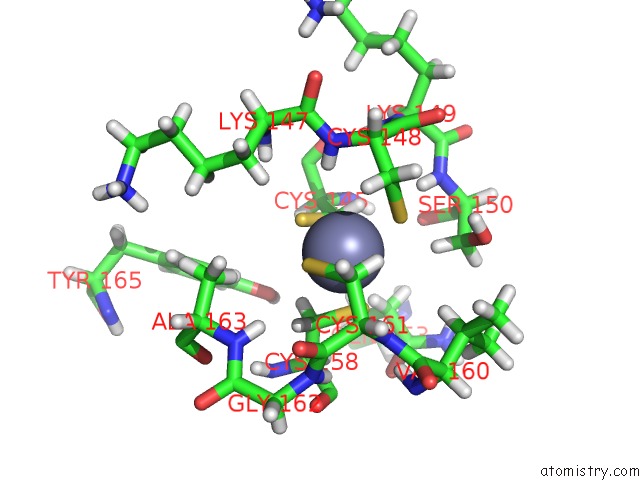
Mono view
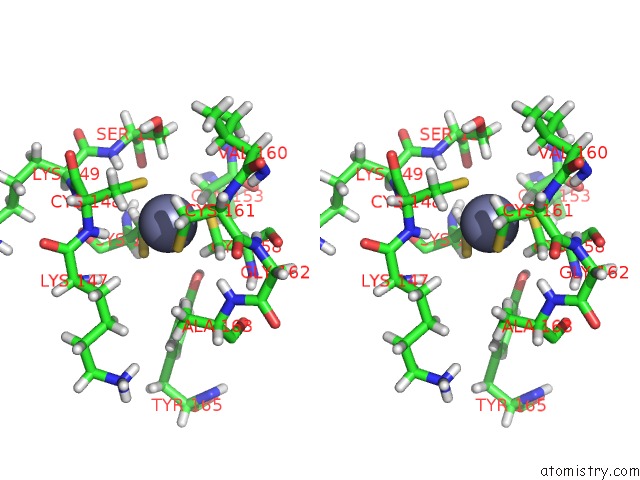
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Structure of the Escherichia Coli Methionyl-Trna Synthetase Complexed with Beta-Methionine within 5.0Å range:
|
Reference:
G.Nigro,
S.Bourcier,
C.Lazennec-Schurdevin,
E.Schmitt,
P.Marliere,
Y.Mechulam.
Use of BETA3-Methionine As An Amino Acid Substrate of Escherichia Coli Methionyl-Trna Synthetase. J.Struct.Biol. 07435 2019.
ISSN: ESSN 1095-8657
PubMed: 31862305
DOI: 10.1016/J.JSB.2019.107435
Page generated: Thu Aug 21 19:45:07 2025
ISSN: ESSN 1095-8657
PubMed: 31862305
DOI: 10.1016/J.JSB.2019.107435
Last articles
Zn in 7AUWZn in 7AUC
Zn in 7ASU
Zn in 7AT1
Zn in 7ATP
Zn in 7ARD
Zn in 7ASJ
Zn in 7ARC
Zn in 7ARU
Zn in 7ARB