Zinc »
PDB 5x6i-5xne »
5xn8 »
Zinc in PDB 5xn8: Structure of Glycerol Dehydrogenase Crystallised As A Contaminant
Protein crystallography data
The structure of Structure of Glycerol Dehydrogenase Crystallised As A Contaminant, PDB code: 5xn8
was solved by
K.Hatti,
Y.K.Mathiharan,
N.Srinivasan,
M.R.N.Murthy,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 56.46 / 2.33 |
| Space group | P 4 |
| Cell size a, b, c (Å), α, β, γ (°) | 178.550, 178.550, 80.090, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 23.6 / 25.1 |
Zinc Binding Sites:
The binding sites of Zinc atom in the Structure of Glycerol Dehydrogenase Crystallised As A Contaminant
(pdb code 5xn8). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 4 binding sites of Zinc where determined in the Structure of Glycerol Dehydrogenase Crystallised As A Contaminant, PDB code: 5xn8:
Jump to Zinc binding site number: 1; 2; 3; 4;
In total 4 binding sites of Zinc where determined in the Structure of Glycerol Dehydrogenase Crystallised As A Contaminant, PDB code: 5xn8:
Jump to Zinc binding site number: 1; 2; 3; 4;
Zinc binding site 1 out of 4 in 5xn8
Go back to
Zinc binding site 1 out
of 4 in the Structure of Glycerol Dehydrogenase Crystallised As A Contaminant
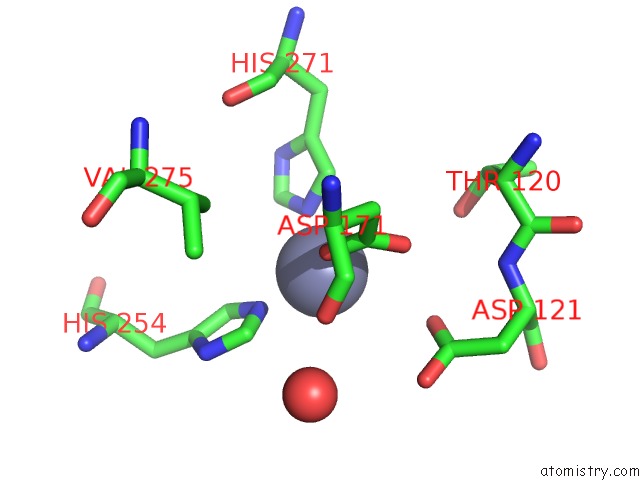
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Structure of Glycerol Dehydrogenase Crystallised As A Contaminant within 5.0Å range:
|
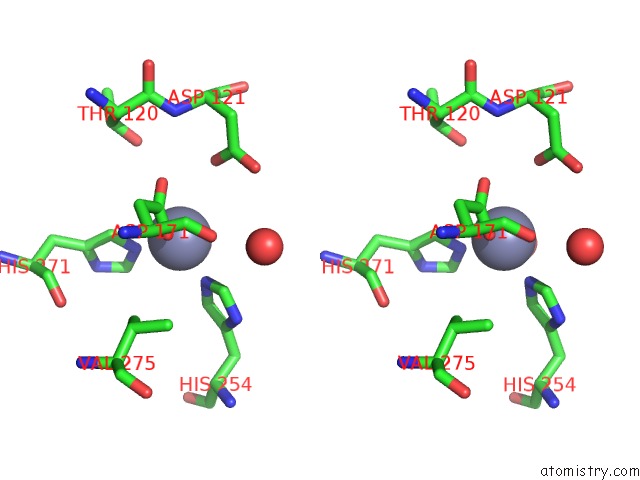
Zinc binding site 2 out of 4 in 5xn8
Go back to
Zinc binding site 2 out
of 4 in the Structure of Glycerol Dehydrogenase Crystallised As A Contaminant

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of Structure of Glycerol Dehydrogenase Crystallised As A Contaminant within 5.0Å range:
|
Zinc binding site 3 out of 4 in 5xn8
Go back to
Zinc binding site 3 out
of 4 in the Structure of Glycerol Dehydrogenase Crystallised As A Contaminant

Mono view
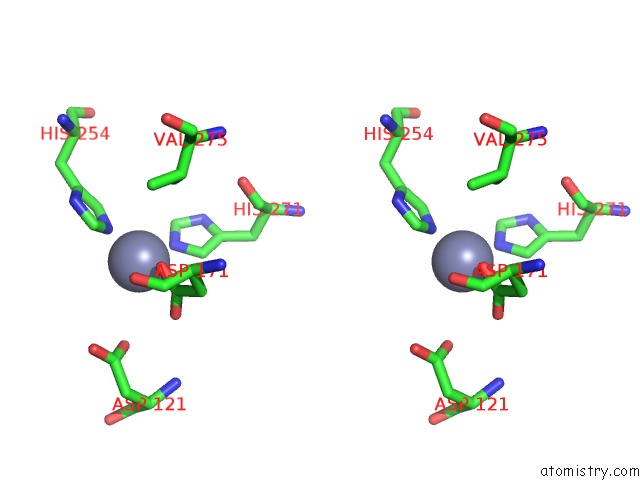
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 3 of Structure of Glycerol Dehydrogenase Crystallised As A Contaminant within 5.0Å range:
|
Zinc binding site 4 out of 4 in 5xn8
Go back to
Zinc binding site 4 out
of 4 in the Structure of Glycerol Dehydrogenase Crystallised As A Contaminant
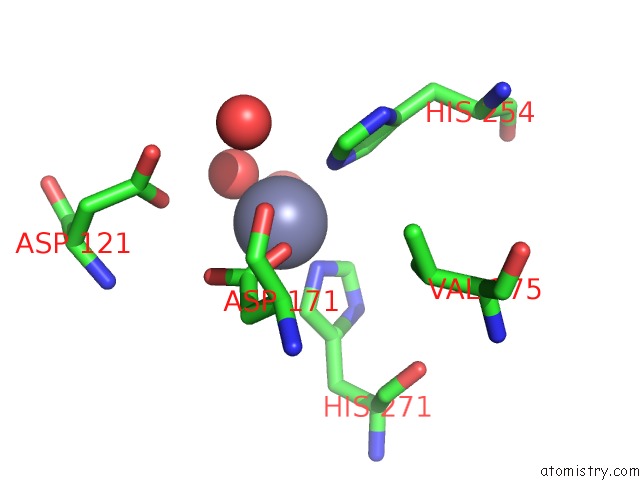
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 4 of Structure of Glycerol Dehydrogenase Crystallised As A Contaminant within 5.0Å range:
|
Reference:
K.Hatti,
Y.K.Mathiharan,
N.Srinivasan,
M.R.N.Murthy.
Seeing But Not Believing: the Structure of Glycerol Dehydrogenase Initially Assumed to Be the Structure of A Survival Protein From Salmonella Typhimurium Acta Crystallogr.,Sect.D V. 73 609 2017.
ISSN: ISSN 0907-4449
DOI: 10.1107/S2059798317007677
Page generated: Mon Oct 28 15:08:58 2024
ISSN: ISSN 0907-4449
DOI: 10.1107/S2059798317007677
Last articles
Mg in 6K9UMg in 6K8B
Mg in 6K85
Mg in 6K89
Mg in 6K86
Mg in 6K7L
Mg in 6K7M
Mg in 6K7N
Mg in 6K82
Mg in 6K61