Zinc »
PDB 5onk-5phh »
5oqj »
Zinc in PDB 5oqj: Structure of Yeast Transcription Pre-Initiation Complex with Tfiih
Enzymatic activity of Structure of Yeast Transcription Pre-Initiation Complex with Tfiih
All present enzymatic activity of Structure of Yeast Transcription Pre-Initiation Complex with Tfiih:
2.7.7.6; 3.6.4.12;
2.7.7.6; 3.6.4.12;
Other elements in 5oqj:
The structure of Structure of Yeast Transcription Pre-Initiation Complex with Tfiih also contains other interesting chemical elements:
| Magnesium | (Mg) | 1 atom |
| Iron | (Fe) | 4 atoms |
Zinc Binding Sites:
Pages:
>>> Page 1 <<< Page 2, Binding sites: 11 - 16;Binding sites:
The binding sites of Zinc atom in the Structure of Yeast Transcription Pre-Initiation Complex with Tfiih (pdb code 5oqj). This binding sites where shown within 5.0 Angstroms radius around Zinc atom.In total 16 binding sites of Zinc where determined in the Structure of Yeast Transcription Pre-Initiation Complex with Tfiih, PDB code: 5oqj:
Jump to Zinc binding site number: 1; 2; 3; 4; 5; 6; 7; 8; 9; 10;
Zinc binding site 1 out of 16 in 5oqj
Go back to
Zinc binding site 1 out
of 16 in the Structure of Yeast Transcription Pre-Initiation Complex with Tfiih
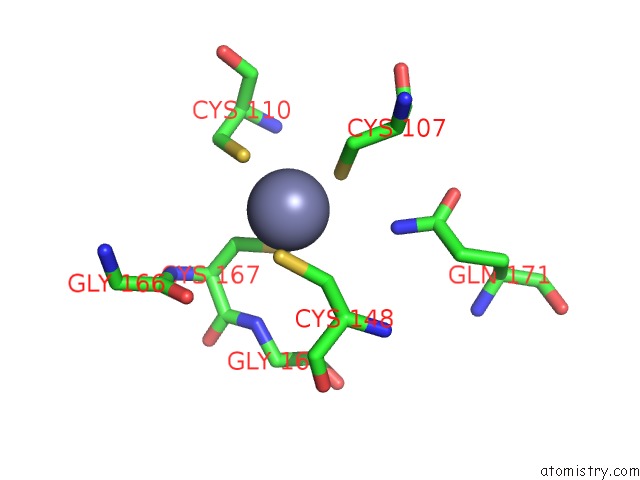
Mono view
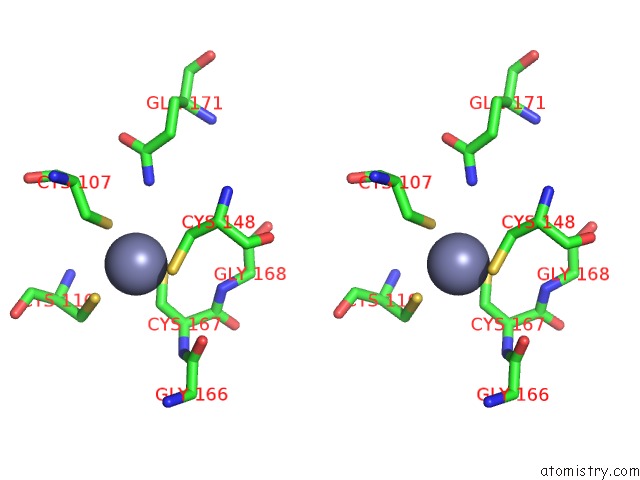
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Structure of Yeast Transcription Pre-Initiation Complex with Tfiih within 5.0Å range:
|
Zinc binding site 2 out of 16 in 5oqj
Go back to
Zinc binding site 2 out
of 16 in the Structure of Yeast Transcription Pre-Initiation Complex with Tfiih
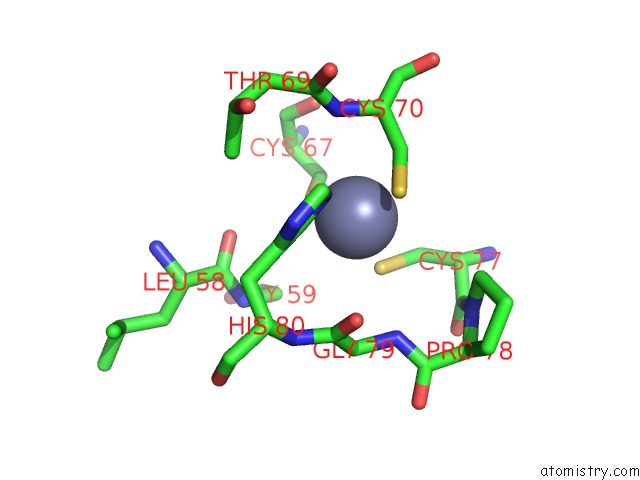
Mono view
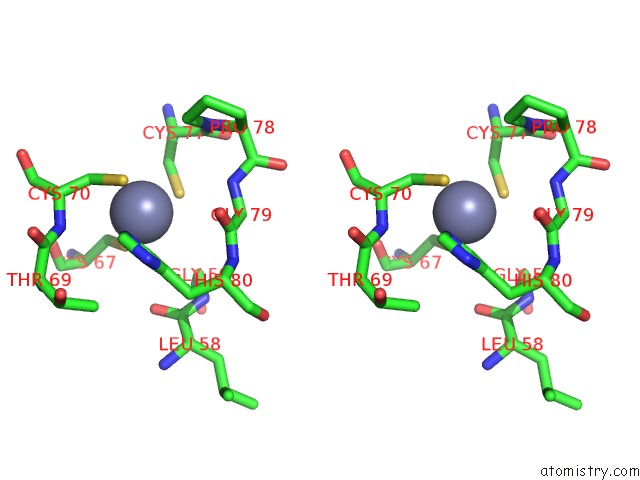
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of Structure of Yeast Transcription Pre-Initiation Complex with Tfiih within 5.0Å range:
|
Zinc binding site 3 out of 16 in 5oqj
Go back to
Zinc binding site 3 out
of 16 in the Structure of Yeast Transcription Pre-Initiation Complex with Tfiih
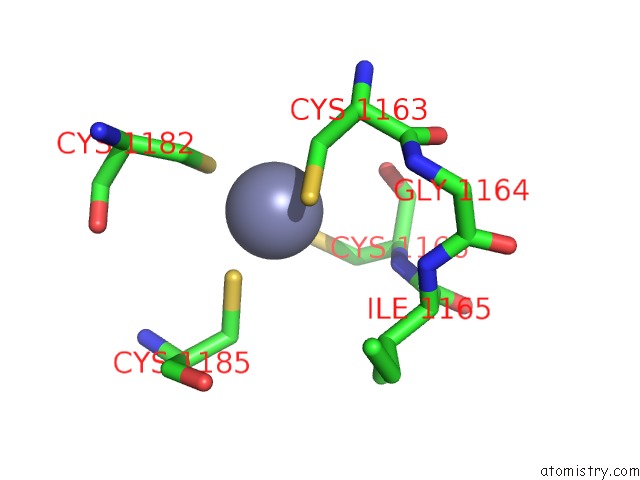
Mono view
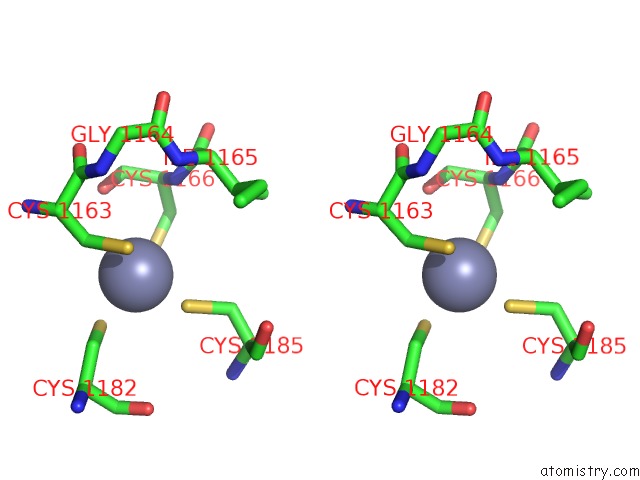
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 3 of Structure of Yeast Transcription Pre-Initiation Complex with Tfiih within 5.0Å range:
|
Zinc binding site 4 out of 16 in 5oqj
Go back to
Zinc binding site 4 out
of 16 in the Structure of Yeast Transcription Pre-Initiation Complex with Tfiih
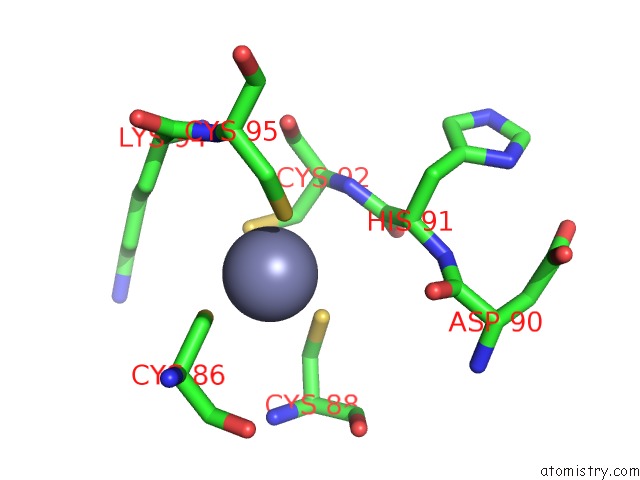
Mono view
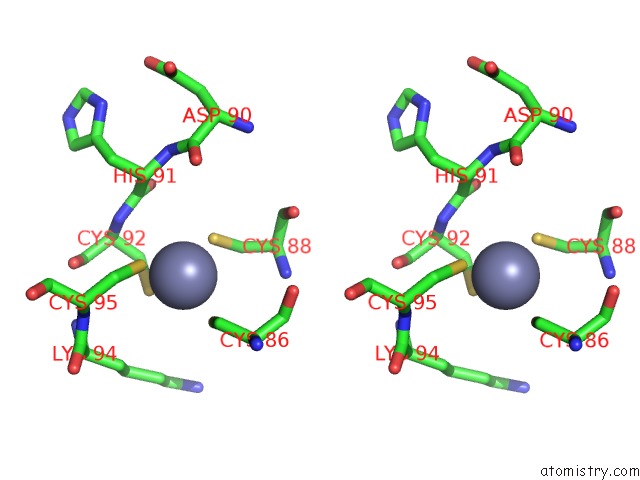
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 4 of Structure of Yeast Transcription Pre-Initiation Complex with Tfiih within 5.0Å range:
|
Zinc binding site 5 out of 16 in 5oqj
Go back to
Zinc binding site 5 out
of 16 in the Structure of Yeast Transcription Pre-Initiation Complex with Tfiih
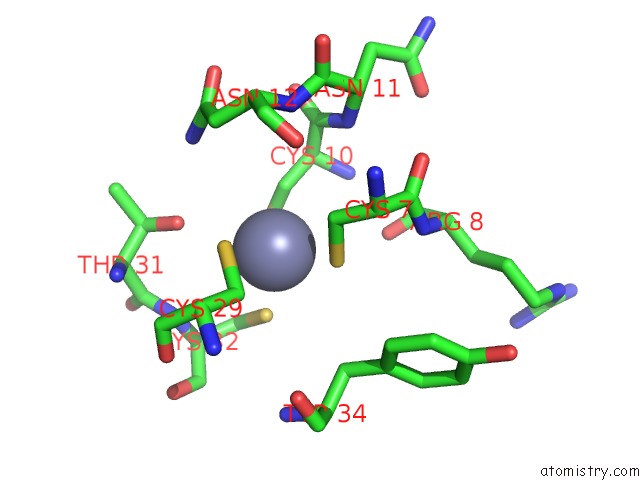
Mono view
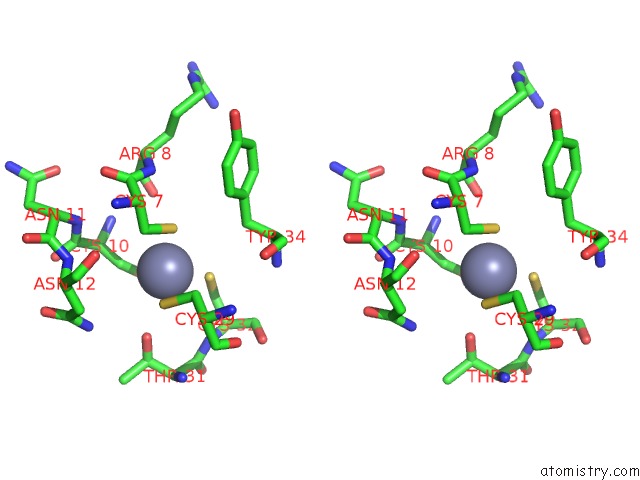
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 5 of Structure of Yeast Transcription Pre-Initiation Complex with Tfiih within 5.0Å range:
|
Zinc binding site 6 out of 16 in 5oqj
Go back to
Zinc binding site 6 out
of 16 in the Structure of Yeast Transcription Pre-Initiation Complex with Tfiih
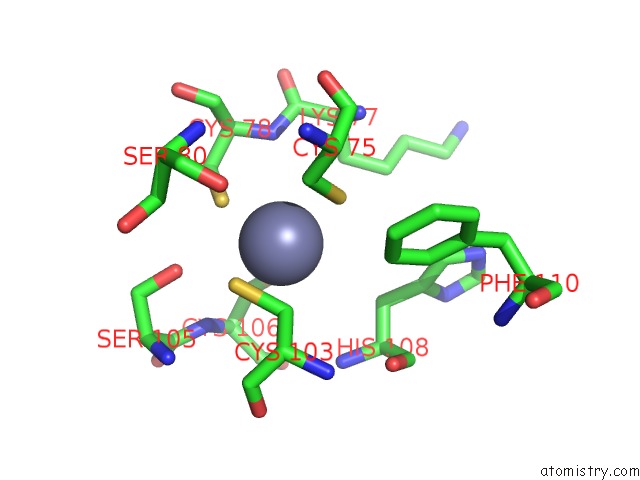
Mono view
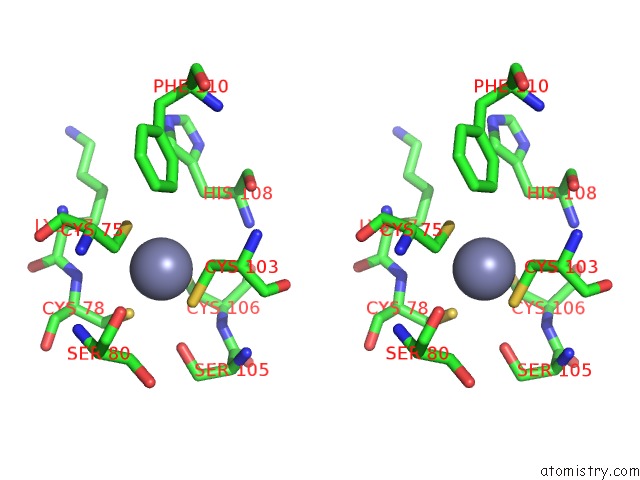
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 6 of Structure of Yeast Transcription Pre-Initiation Complex with Tfiih within 5.0Å range:
|
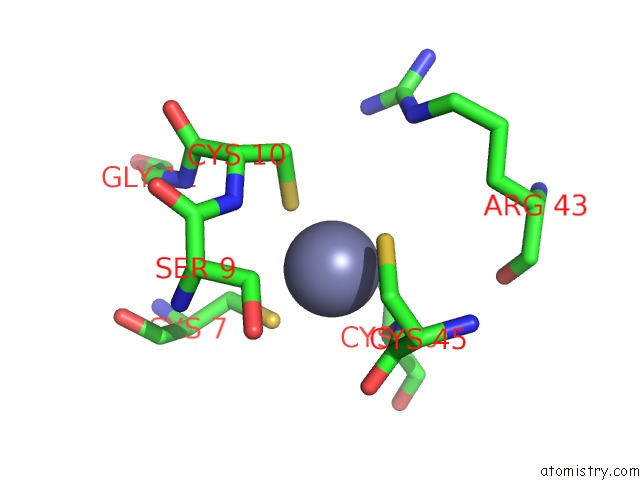
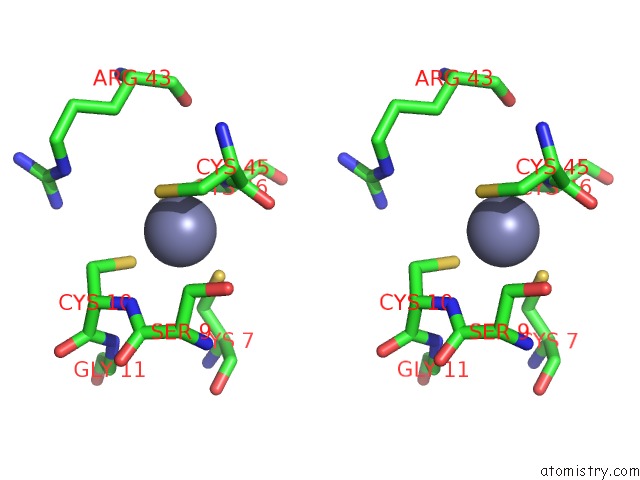
Zinc binding site 7 out of 16 in 5oqj
Go back to
Zinc binding site 7 out
of 16 in the Structure of Yeast Transcription Pre-Initiation Complex with Tfiih
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 7 of Structure of Yeast Transcription Pre-Initiation Complex with Tfiih within 5.0Å range:
|
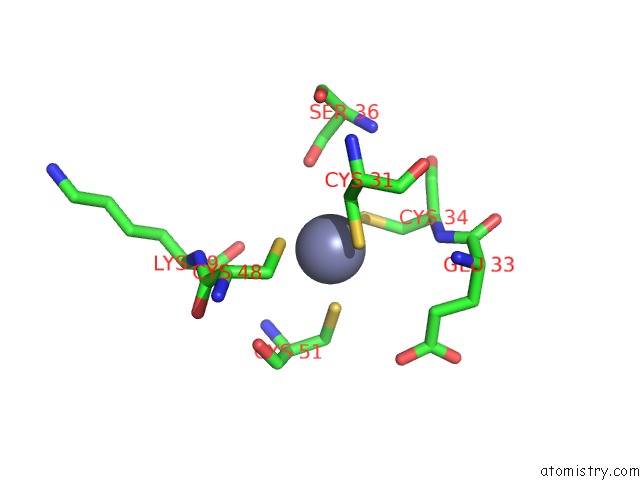
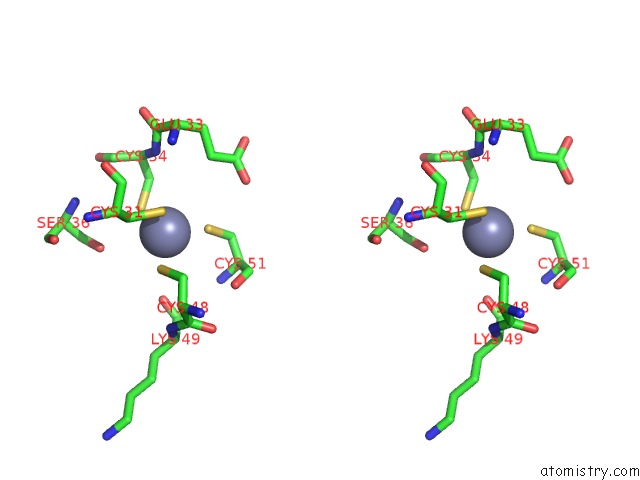
Zinc binding site 8 out of 16 in 5oqj
Go back to
Zinc binding site 8 out
of 16 in the Structure of Yeast Transcription Pre-Initiation Complex with Tfiih
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 8 of Structure of Yeast Transcription Pre-Initiation Complex with Tfiih within 5.0Å range:
|
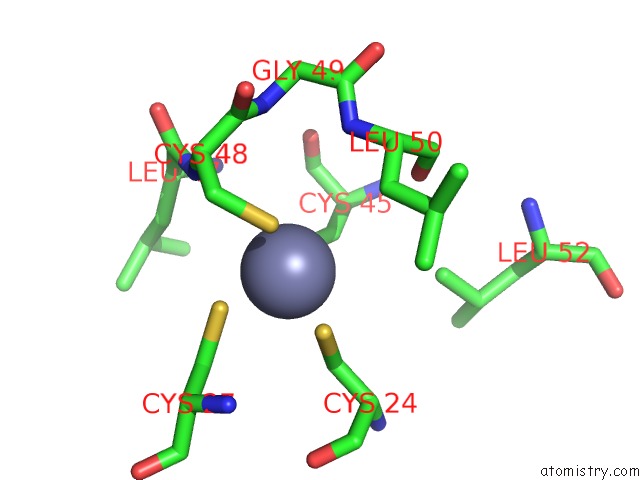
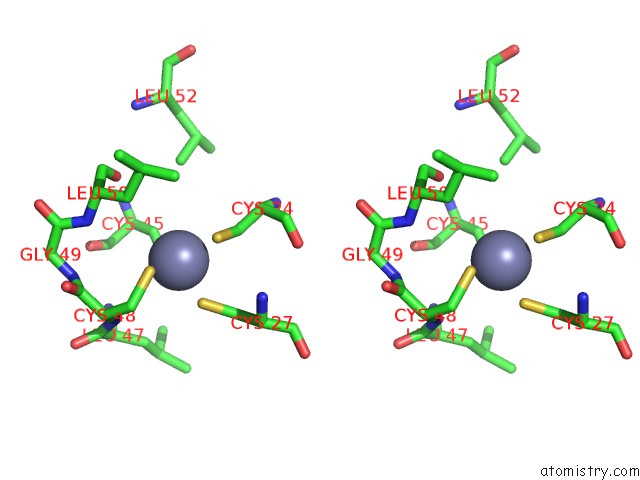
Zinc binding site 9 out of 16 in 5oqj
Go back to
Zinc binding site 9 out
of 16 in the Structure of Yeast Transcription Pre-Initiation Complex with Tfiih
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 9 of Structure of Yeast Transcription Pre-Initiation Complex with Tfiih within 5.0Å range:
|
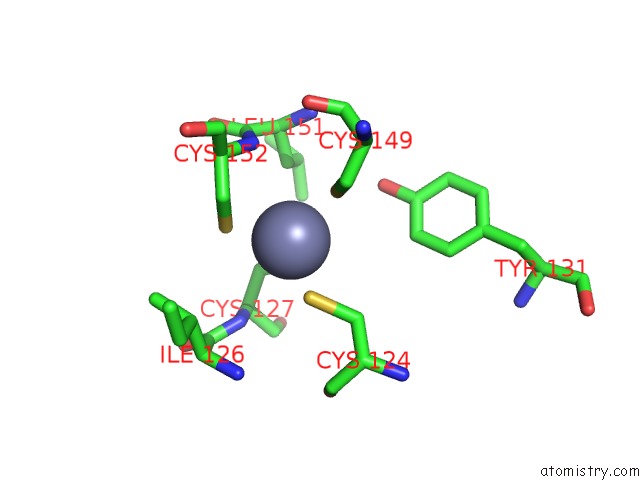
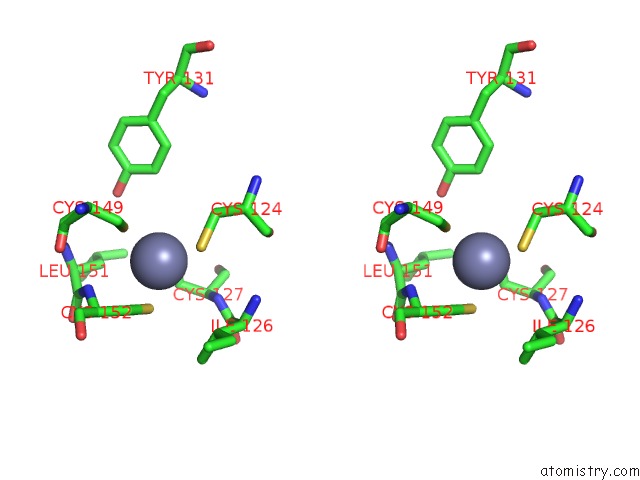
Zinc binding site 10 out of 16 in 5oqj
Go back to
Zinc binding site 10 out
of 16 in the Structure of Yeast Transcription Pre-Initiation Complex with Tfiih
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 10 of Structure of Yeast Transcription Pre-Initiation Complex with Tfiih within 5.0Å range:
|
Reference:
S.Schilbach,
M.Hantsche,
D.Tegunov,
C.Dienemann,
C.Wigge,
H.Urlaub,
P.Cramer.
Structures of Transcription Pre-Initiation Complex with Tfiih and Mediator. Nature V. 551 204 2017.
ISSN: ESSN 1476-4687
PubMed: 29088706
DOI: 10.1038/NATURE24282
Page generated: Sun Oct 27 23:46:47 2024
ISSN: ESSN 1476-4687
PubMed: 29088706
DOI: 10.1038/NATURE24282
Last articles
Mg in 5CZ2Mg in 5CYP
Mg in 5CYV
Mg in 5CYO
Mg in 5CYR
Mg in 5CX7
Mg in 5CVC
Mg in 5CX6
Mg in 5CUY
Mg in 5CVW