Zinc »
PDB 5dag-5drq »
5dpe »
Zinc in PDB 5dpe: Thermolysin in Complex with Inhibitor.
Enzymatic activity of Thermolysin in Complex with Inhibitor.
All present enzymatic activity of Thermolysin in Complex with Inhibitor.:
3.4.24.27;
3.4.24.27;
Protein crystallography data
The structure of Thermolysin in Complex with Inhibitor., PDB code: 5dpe
was solved by
S.G.Krimmer,
A.Heine,
G.Klebe,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 34.17 / 1.34 |
| Space group | P 61 2 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 92.690, 92.690, 130.261, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 12.7 / 15.6 |
Other elements in 5dpe:
The structure of Thermolysin in Complex with Inhibitor. also contains other interesting chemical elements:
| Calcium | (Ca) | 4 atoms |
Zinc Binding Sites:
The binding sites of Zinc atom in the Thermolysin in Complex with Inhibitor.
(pdb code 5dpe). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total only one binding site of Zinc was determined in the Thermolysin in Complex with Inhibitor., PDB code: 5dpe:
In total only one binding site of Zinc was determined in the Thermolysin in Complex with Inhibitor., PDB code: 5dpe:
Zinc binding site 1 out of 1 in 5dpe
Go back to
Zinc binding site 1 out
of 1 in the Thermolysin in Complex with Inhibitor.
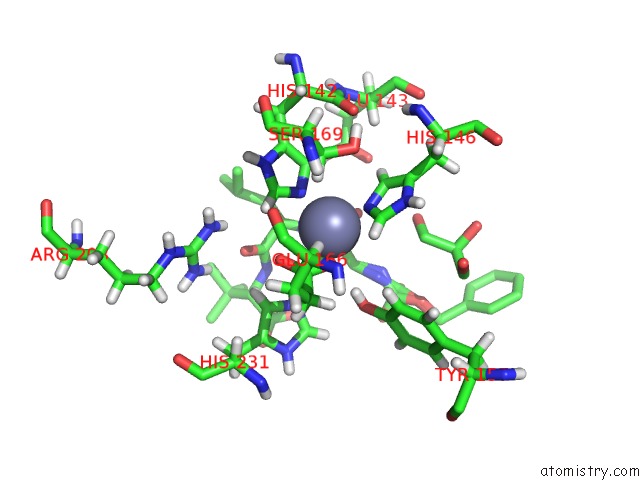
Mono view
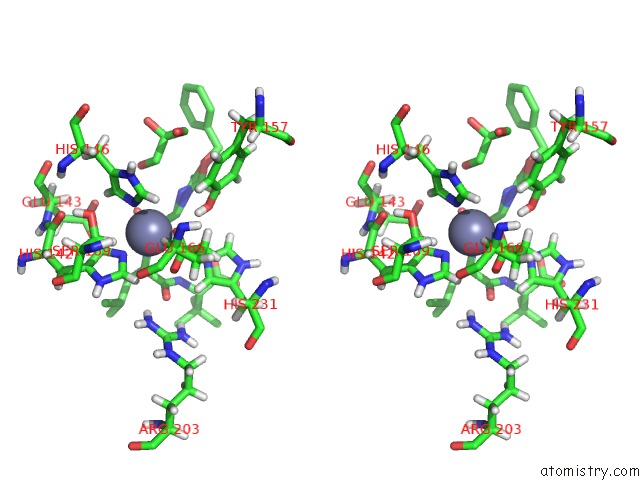
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Thermolysin in Complex with Inhibitor. within 5.0Å range:
|
Reference:
S.G.Krimmer,
G.Klebe.
Thermodynamics of Protein-Ligand Interactions As A Reference For Computational Analysis: How to Assess Accuracy, Reliability and Relevance of Experimental Data. J. Comput. Aided Mol. Des. V. 29 867 2015.
ISSN: ISSN 1573-4951
PubMed: 26376645
DOI: 10.1007/S10822-015-9867-Y
Page generated: Sun Oct 27 14:53:28 2024
ISSN: ISSN 1573-4951
PubMed: 26376645
DOI: 10.1007/S10822-015-9867-Y
Last articles
Mg in 4ZESMg in 4ZCW
Mg in 4ZDQ
Mg in 4ZDK
Mg in 4ZD6
Mg in 4ZDJ
Mg in 4ZCM
Mg in 4Z71
Mg in 4ZA0
Mg in 4ZCL