Zinc »
PDB 4jsh-4k4t »
4k25 »
Zinc in PDB 4k25: Crystal Structure of Yeast QRI7 Homodimer
Protein crystallography data
The structure of Crystal Structure of Yeast QRI7 Homodimer, PDB code: 4k25
was solved by
D.Neculai,
L.Wan,
D.Y.Mao,
F.Sicheri,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 20.58 / 2.88 |
| Space group | P 21 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 64.597, 130.983, 47.866, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 24 / 29.5 |
Other elements in 4k25:
The structure of Crystal Structure of Yeast QRI7 Homodimer also contains other interesting chemical elements:
| Calcium | (Ca) | 1 atom |
Zinc Binding Sites:
The binding sites of Zinc atom in the Crystal Structure of Yeast QRI7 Homodimer
(pdb code 4k25). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total only one binding site of Zinc was determined in the Crystal Structure of Yeast QRI7 Homodimer, PDB code: 4k25:
In total only one binding site of Zinc was determined in the Crystal Structure of Yeast QRI7 Homodimer, PDB code: 4k25:
Zinc binding site 1 out of 1 in 4k25
Go back to
Zinc binding site 1 out
of 1 in the Crystal Structure of Yeast QRI7 Homodimer

Mono view
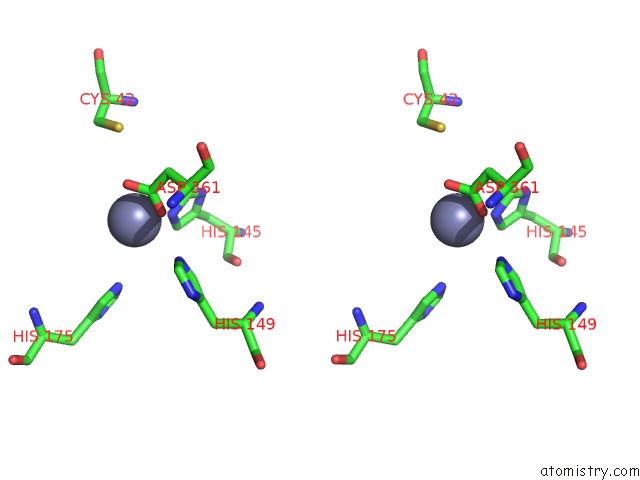
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Crystal Structure of Yeast QRI7 Homodimer within 5.0Å range:
|
Reference:
L.C.Wan,
D.Y.Mao,
D.Neculai,
J.Strecker,
D.Chiovitti,
I.Kurinov,
G.Poda,
N.Thevakumaran,
F.Yuan,
R.K.Szilard,
E.Lissina,
C.Nislow,
A.A.Caudy,
D.Durocher,
F.Sicheri.
Reconstitution and Characterization of Eukaryotic N6-Threonylcarbamoylation of Trna Using A Minimal Enzyme System. Nucleic Acids Res. V. 41 6332 2013.
ISSN: ISSN 0305-1048
PubMed: 23620299
DOI: 10.1093/NAR/GKT322
Page generated: Wed Aug 20 19:21:23 2025
ISSN: ISSN 0305-1048
PubMed: 23620299
DOI: 10.1093/NAR/GKT322
Last articles
Zn in 5C0KZn in 5BZ0
Zn in 5BWZ
Zn in 5BYI
Zn in 5BWK
Zn in 5BWO
Zn in 5BWN
Zn in 5BWL
Zn in 5BUO
Zn in 5BVR