Zinc »
PDB 2y7g-2yqp »
2ypa »
Zinc in PDB 2ypa: Structure of the Scl:E47:LMO2:LDB1 Complex Bound to Dna
Protein crystallography data
The structure of Structure of the Scl:E47:LMO2:LDB1 Complex Bound to Dna, PDB code: 2ypa
was solved by
K.El Omari,
S.J.Hoosdally,
K.Tuladhar,
D.Karia,
E.Ponsele,
O.Platonova,
P.Vyas,
R.Patient,
C.Porcher,
E.J.Mancini,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 30.15 / 2.80 |
| Space group | F 2 2 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 102.966, 141.044, 148.793, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 22.401 / 26.945 |
Zinc Binding Sites:
The binding sites of Zinc atom in the Structure of the Scl:E47:LMO2:LDB1 Complex Bound to Dna
(pdb code 2ypa). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 4 binding sites of Zinc where determined in the Structure of the Scl:E47:LMO2:LDB1 Complex Bound to Dna, PDB code: 2ypa:
Jump to Zinc binding site number: 1; 2; 3; 4;
In total 4 binding sites of Zinc where determined in the Structure of the Scl:E47:LMO2:LDB1 Complex Bound to Dna, PDB code: 2ypa:
Jump to Zinc binding site number: 1; 2; 3; 4;
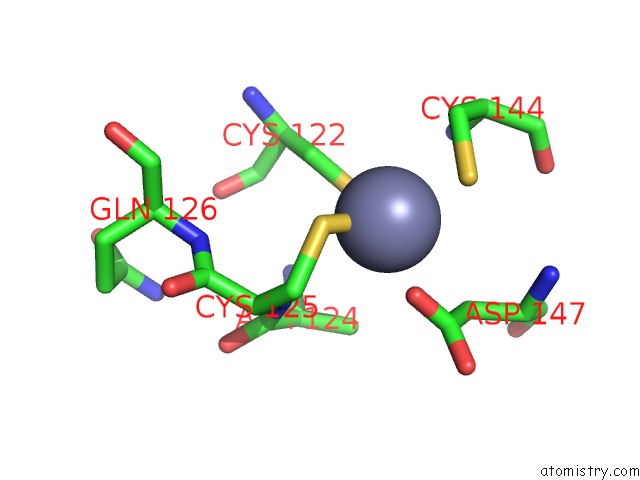
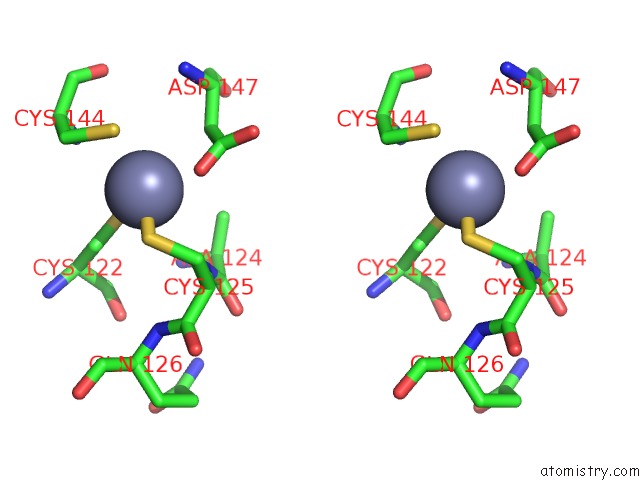
Zinc binding site 1 out of 4 in 2ypa
Go back to
Zinc binding site 1 out
of 4 in the Structure of the Scl:E47:LMO2:LDB1 Complex Bound to Dna
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Structure of the Scl:E47:LMO2:LDB1 Complex Bound to Dna within 5.0Å range:
|
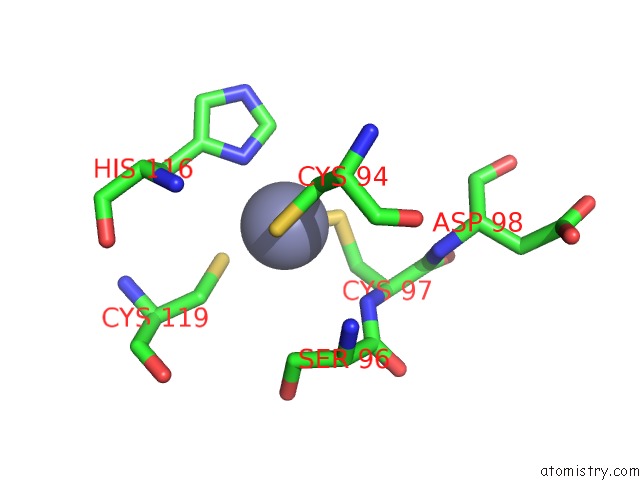
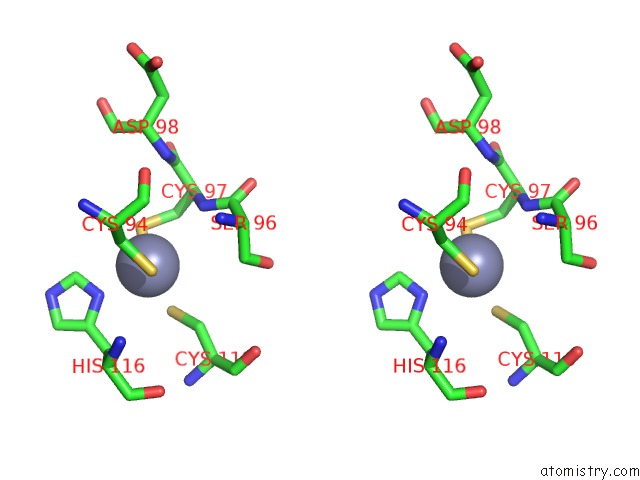
Zinc binding site 2 out of 4 in 2ypa
Go back to
Zinc binding site 2 out
of 4 in the Structure of the Scl:E47:LMO2:LDB1 Complex Bound to Dna
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of Structure of the Scl:E47:LMO2:LDB1 Complex Bound to Dna within 5.0Å range:
|
Zinc binding site 3 out of 4 in 2ypa
Go back to
Zinc binding site 3 out
of 4 in the Structure of the Scl:E47:LMO2:LDB1 Complex Bound to Dna
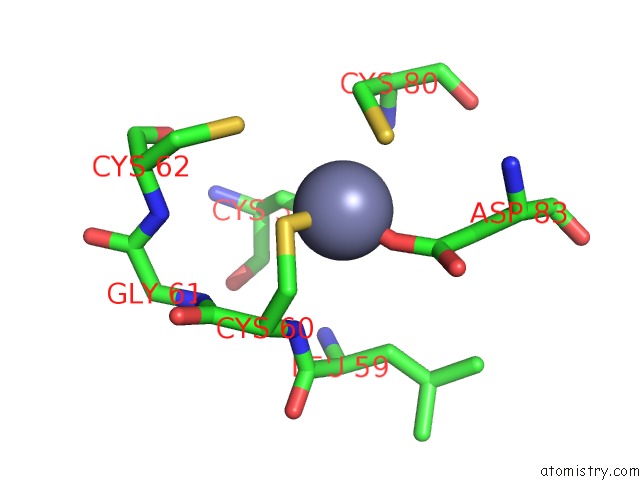
Mono view
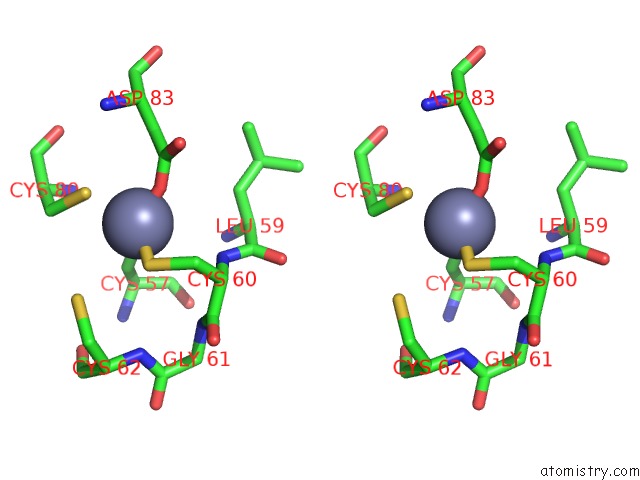
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 3 of Structure of the Scl:E47:LMO2:LDB1 Complex Bound to Dna within 5.0Å range:
|
Zinc binding site 4 out of 4 in 2ypa
Go back to
Zinc binding site 4 out
of 4 in the Structure of the Scl:E47:LMO2:LDB1 Complex Bound to Dna
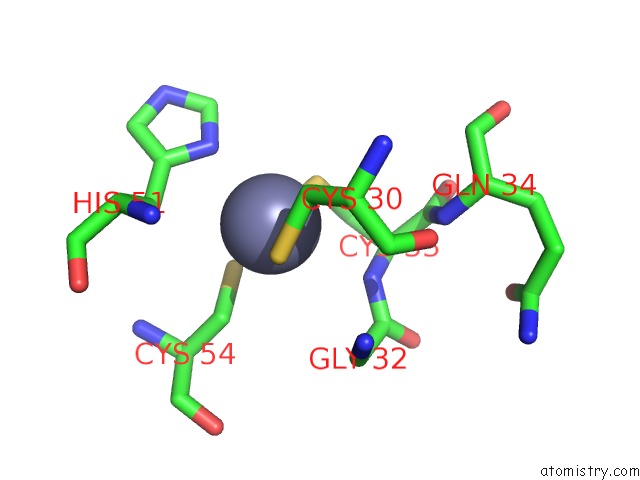
Mono view
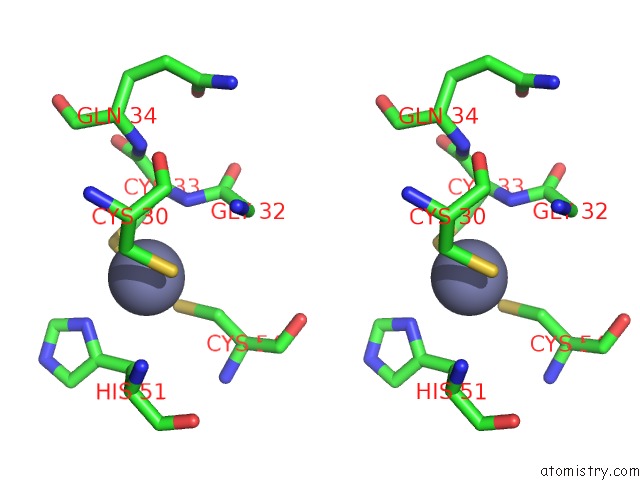
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 4 of Structure of the Scl:E47:LMO2:LDB1 Complex Bound to Dna within 5.0Å range:
|
Reference:
K.El Omari,
S.J.Hoosdally,
K.Tuladhar,
D.Karia,
E.Hall-Ponsele,
O.Platonova,
P.Vyas,
R.Patient,
C.Porcher,
E.J.Mancini.
Structural Basis For LMO2-Driven Recruitment of the Scl:E47BHLH Heterodimer to Hematopoietic-Specific Transcriptional Targets. Cell Rep. V. 4 135 2013.
ISSN: ISSN 2211-1247
PubMed: 23831025
DOI: 10.1016/J.CELREP.2013.06.008
Page generated: Thu Oct 17 05:54:22 2024
ISSN: ISSN 2211-1247
PubMed: 23831025
DOI: 10.1016/J.CELREP.2013.06.008
Last articles
F in 5UZ0F in 5UY8
F in 5UX4
F in 5UV3
F in 5UWF
F in 5UVF
F in 5UVW
F in 5UWD
F in 5UW0
F in 5UU3