Zinc »
PDB 1y8q-1ykf »
1ybq »
Zinc in PDB 1ybq: Crystal Structure of Escherichia Coli Isoaspartyl Dipeptidase Mutant D285N Complexed with Beta-Aspartylhistidine
Protein crystallography data
The structure of Crystal Structure of Escherichia Coli Isoaspartyl Dipeptidase Mutant D285N Complexed with Beta-Aspartylhistidine, PDB code: 1ybq
was solved by
R.Marti-Arbona,
V.Fresquet,
J.B.Thoden,
M.L.Davis,
H.M.Holden,
F.M.Raushel,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 30.00 / 2.00 |
| Space group | P 4 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 119.300, 119.300, 138.300, 90.00, 90.00, 90.00 |
| R / Rfree (%) | n/a / n/a |
Zinc Binding Sites:
The binding sites of Zinc atom in the Crystal Structure of Escherichia Coli Isoaspartyl Dipeptidase Mutant D285N Complexed with Beta-Aspartylhistidine
(pdb code 1ybq). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 4 binding sites of Zinc where determined in the Crystal Structure of Escherichia Coli Isoaspartyl Dipeptidase Mutant D285N Complexed with Beta-Aspartylhistidine, PDB code: 1ybq:
Jump to Zinc binding site number: 1; 2; 3; 4;
In total 4 binding sites of Zinc where determined in the Crystal Structure of Escherichia Coli Isoaspartyl Dipeptidase Mutant D285N Complexed with Beta-Aspartylhistidine, PDB code: 1ybq:
Jump to Zinc binding site number: 1; 2; 3; 4;
Zinc binding site 1 out of 4 in 1ybq
Go back to
Zinc binding site 1 out
of 4 in the Crystal Structure of Escherichia Coli Isoaspartyl Dipeptidase Mutant D285N Complexed with Beta-Aspartylhistidine

Mono view
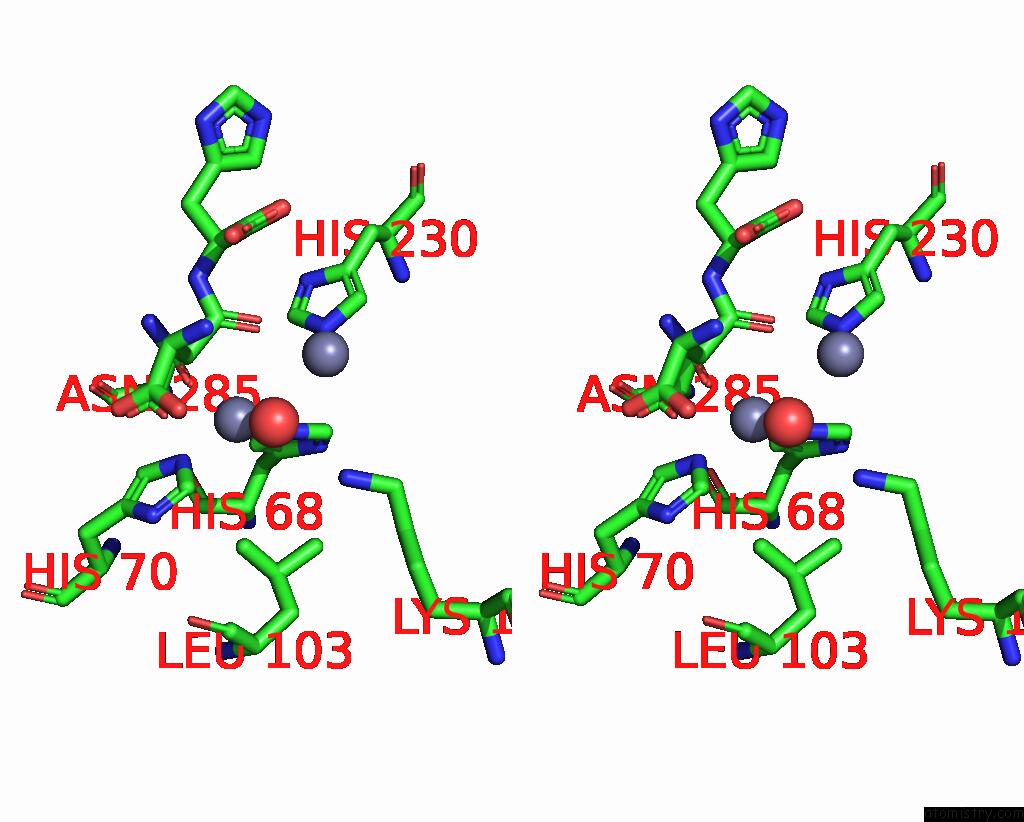
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Crystal Structure of Escherichia Coli Isoaspartyl Dipeptidase Mutant D285N Complexed with Beta-Aspartylhistidine within 5.0Å range:
|
Zinc binding site 2 out of 4 in 1ybq
Go back to
Zinc binding site 2 out
of 4 in the Crystal Structure of Escherichia Coli Isoaspartyl Dipeptidase Mutant D285N Complexed with Beta-Aspartylhistidine

Mono view
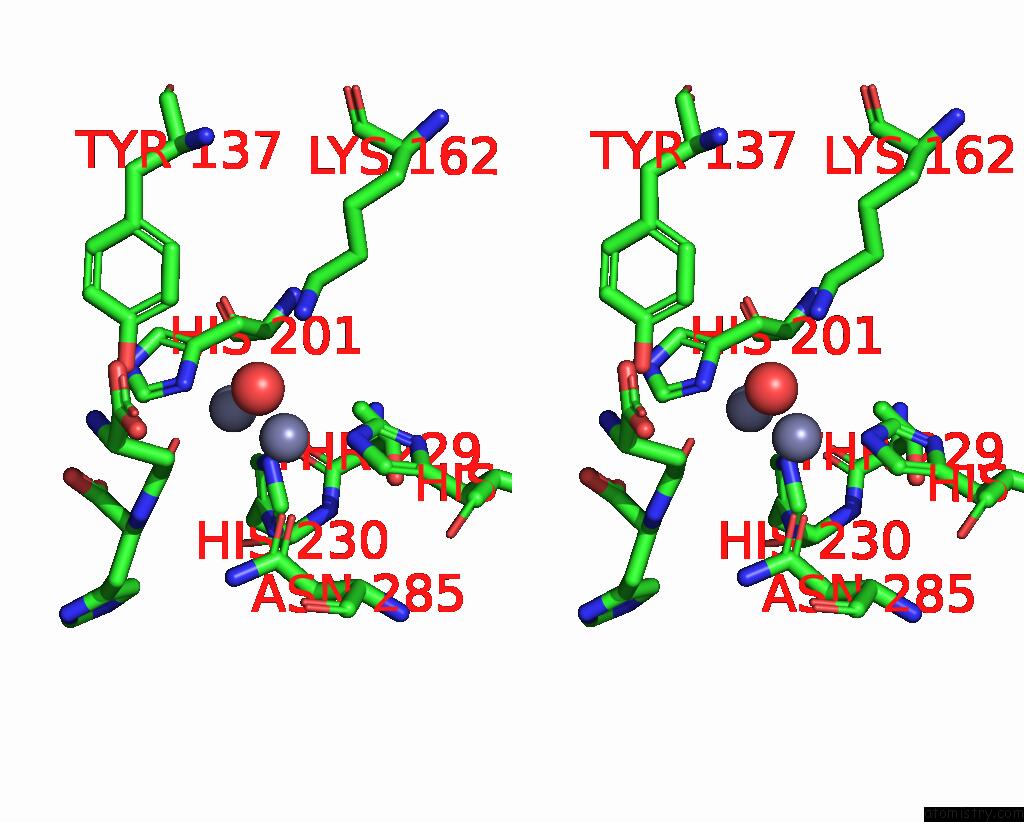
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of Crystal Structure of Escherichia Coli Isoaspartyl Dipeptidase Mutant D285N Complexed with Beta-Aspartylhistidine within 5.0Å range:
|
Zinc binding site 3 out of 4 in 1ybq
Go back to
Zinc binding site 3 out
of 4 in the Crystal Structure of Escherichia Coli Isoaspartyl Dipeptidase Mutant D285N Complexed with Beta-Aspartylhistidine

Mono view
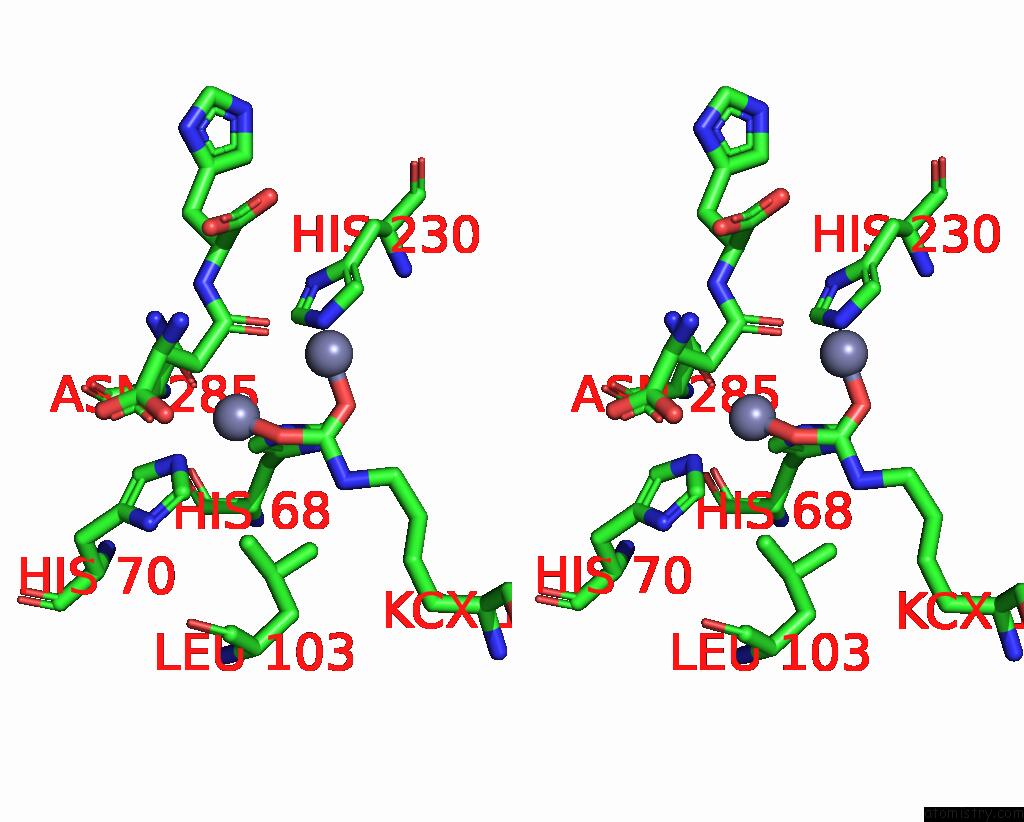
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 3 of Crystal Structure of Escherichia Coli Isoaspartyl Dipeptidase Mutant D285N Complexed with Beta-Aspartylhistidine within 5.0Å range:
|
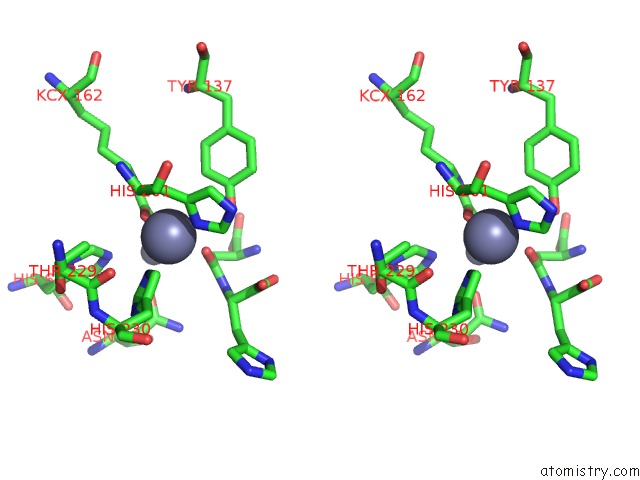
Zinc binding site 4 out of 4 in 1ybq
Go back to
Zinc binding site 4 out
of 4 in the Crystal Structure of Escherichia Coli Isoaspartyl Dipeptidase Mutant D285N Complexed with Beta-Aspartylhistidine
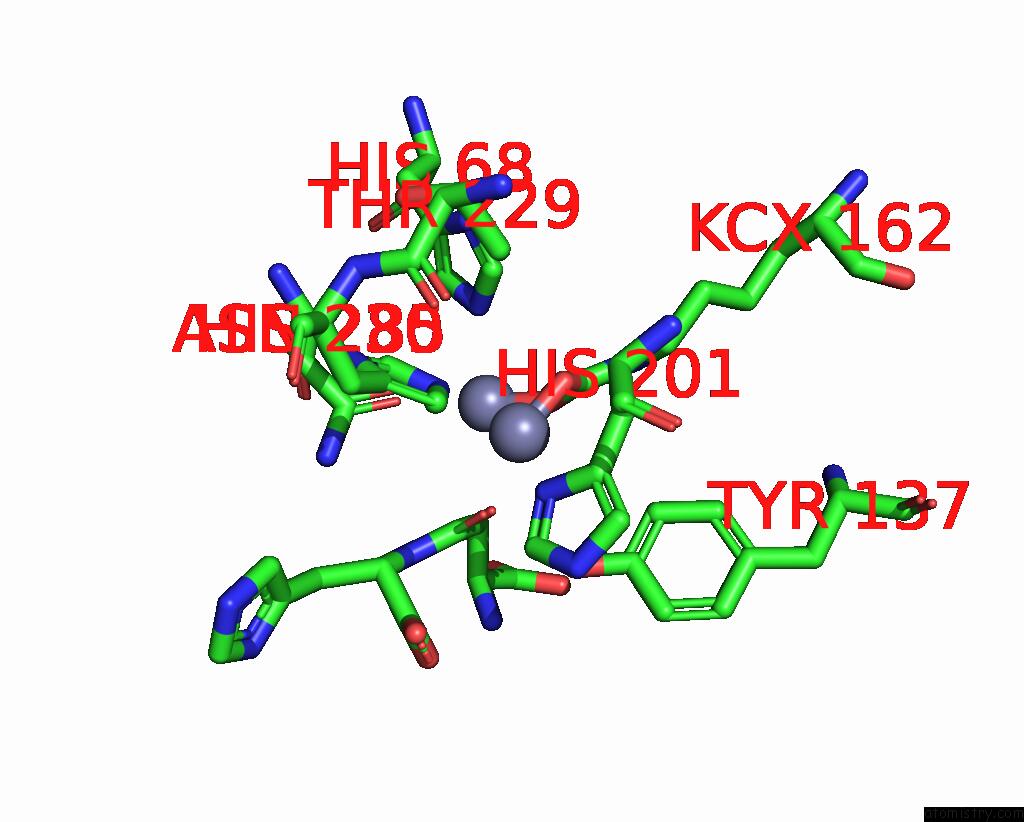
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 4 of Crystal Structure of Escherichia Coli Isoaspartyl Dipeptidase Mutant D285N Complexed with Beta-Aspartylhistidine within 5.0Å range:
|
Reference:
R.Marti-Arbona,
V.Fresquet,
J.B.Thoden,
M.L.Davis,
H.M.Holden,
F.M.Raushel.
Mechanism of the Reaction Catalyzed By Isoaspartyl Dipeptidase From Escherichia Coli. Biochemistry V. 44 7115 2005.
ISSN: ISSN 0006-2960
PubMed: 15882050
DOI: 10.1021/BI050008R
Page generated: Wed Oct 16 20:49:31 2024
ISSN: ISSN 0006-2960
PubMed: 15882050
DOI: 10.1021/BI050008R
Last articles
Mn in 4R43Mn in 4QSH
Mn in 4QSK
Mn in 4QTG
Mn in 4QNK
Mn in 4QRO
Mn in 4QSF
Mn in 4QS5
Mn in 4QRN
Mn in 4QPX