Zinc »
PDB 9iic-9j62 »
9imm »
Zinc in PDB 9imm: Sars-Cov-2 Replication-Transcription Complex Has A Dimer Architecture (Local Drtc) in Post-Capping State
Enzymatic activity of Sars-Cov-2 Replication-Transcription Complex Has A Dimer Architecture (Local Drtc) in Post-Capping State
All present enzymatic activity of Sars-Cov-2 Replication-Transcription Complex Has A Dimer Architecture (Local Drtc) in Post-Capping State:
2.7.7.48; 2.7.7.50; 3.6.4.12; 3.6.4.13;
2.7.7.48; 2.7.7.50; 3.6.4.12; 3.6.4.13;
Zinc Binding Sites:
The binding sites of Zinc atom in the Sars-Cov-2 Replication-Transcription Complex Has A Dimer Architecture (Local Drtc) in Post-Capping State
(pdb code 9imm). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 8 binding sites of Zinc where determined in the Sars-Cov-2 Replication-Transcription Complex Has A Dimer Architecture (Local Drtc) in Post-Capping State, PDB code: 9imm:
Jump to Zinc binding site number: 1; 2; 3; 4; 5; 6; 7; 8;
In total 8 binding sites of Zinc where determined in the Sars-Cov-2 Replication-Transcription Complex Has A Dimer Architecture (Local Drtc) in Post-Capping State, PDB code: 9imm:
Jump to Zinc binding site number: 1; 2; 3; 4; 5; 6; 7; 8;
Zinc binding site 1 out of 8 in 9imm
Go back to
Zinc binding site 1 out
of 8 in the Sars-Cov-2 Replication-Transcription Complex Has A Dimer Architecture (Local Drtc) in Post-Capping State
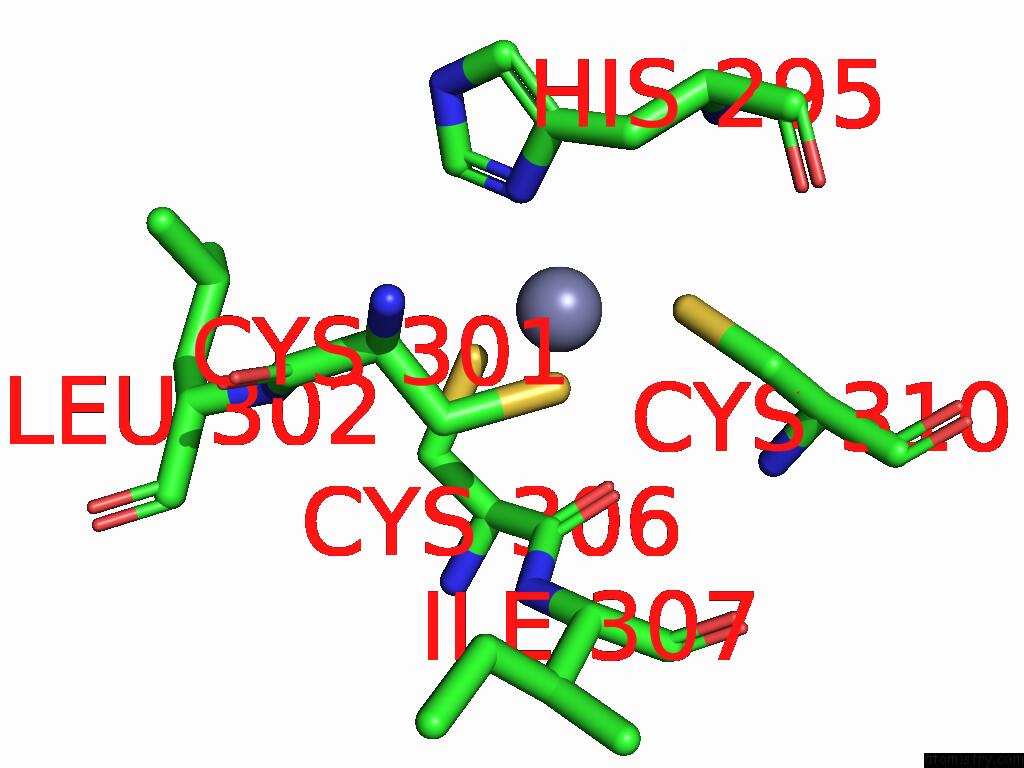
Mono view
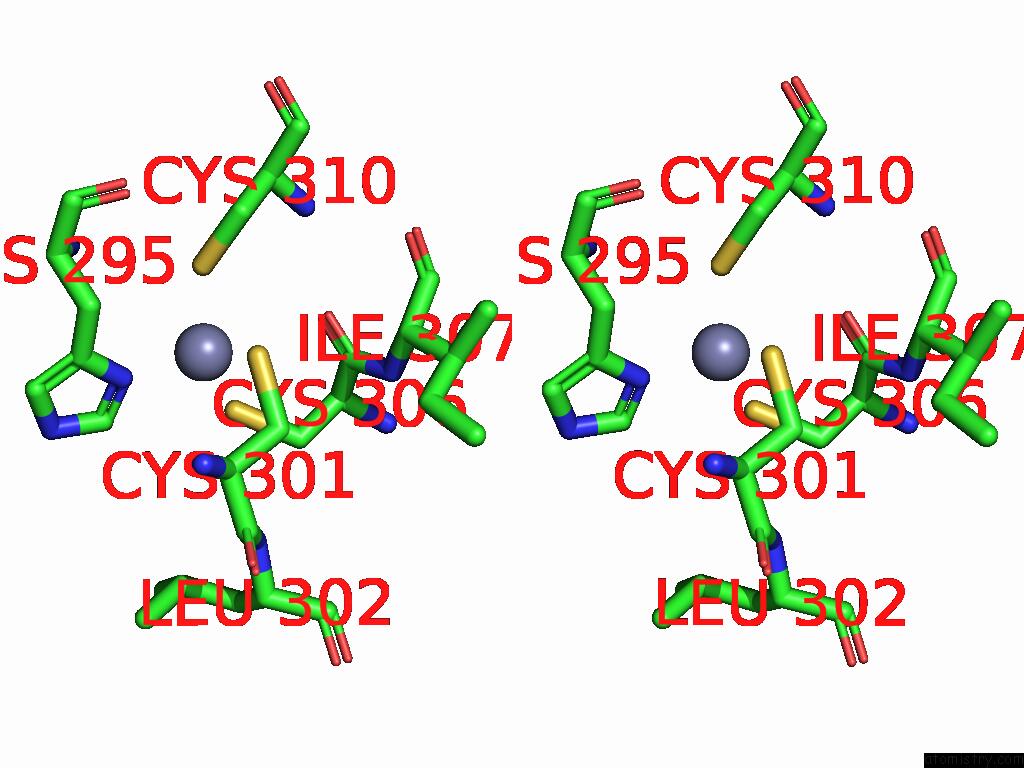
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Sars-Cov-2 Replication-Transcription Complex Has A Dimer Architecture (Local Drtc) in Post-Capping State within 5.0Å range:
|
Zinc binding site 2 out of 8 in 9imm
Go back to
Zinc binding site 2 out
of 8 in the Sars-Cov-2 Replication-Transcription Complex Has A Dimer Architecture (Local Drtc) in Post-Capping State
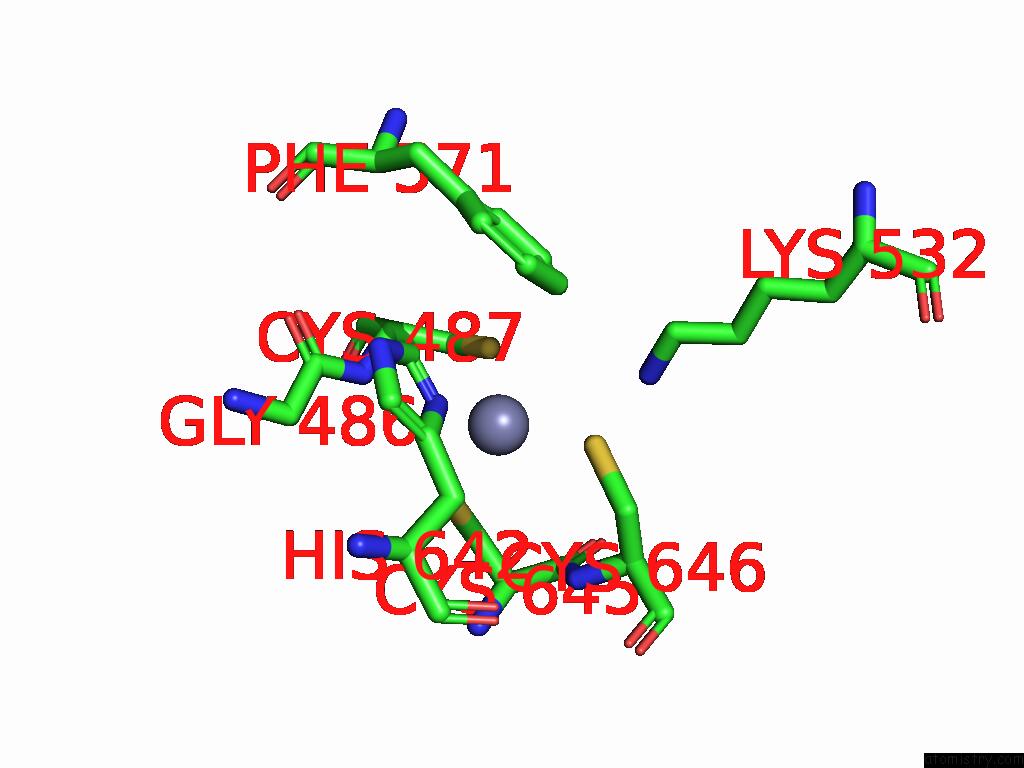
Mono view
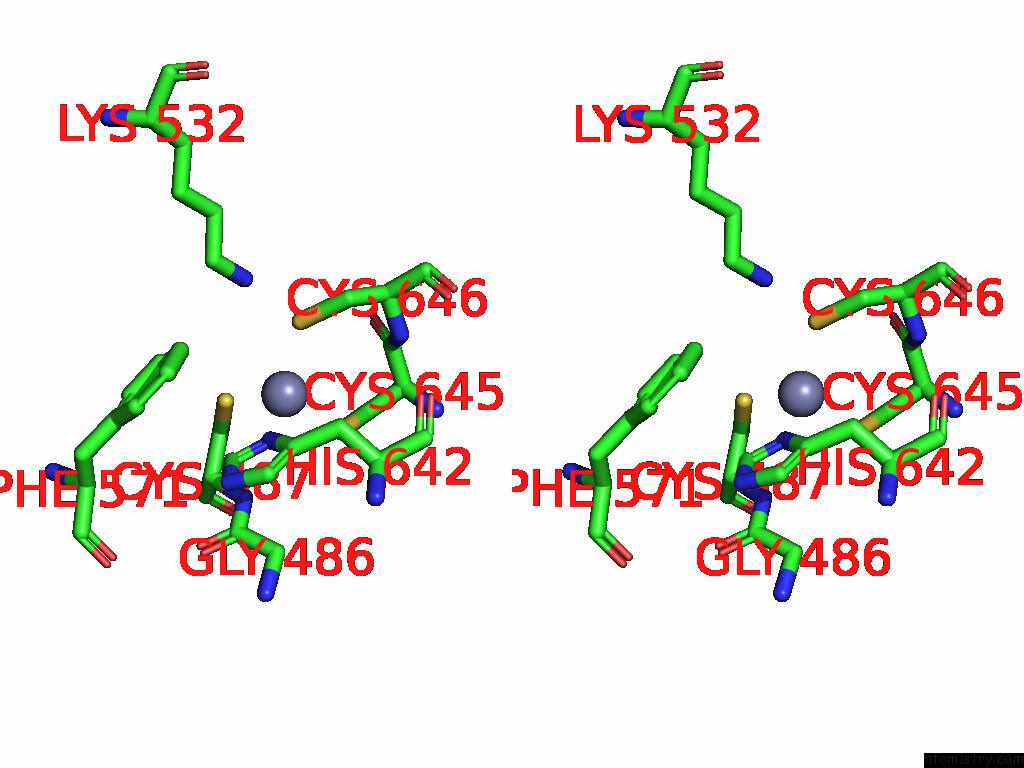
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of Sars-Cov-2 Replication-Transcription Complex Has A Dimer Architecture (Local Drtc) in Post-Capping State within 5.0Å range:
|
Zinc binding site 3 out of 8 in 9imm
Go back to
Zinc binding site 3 out
of 8 in the Sars-Cov-2 Replication-Transcription Complex Has A Dimer Architecture (Local Drtc) in Post-Capping State
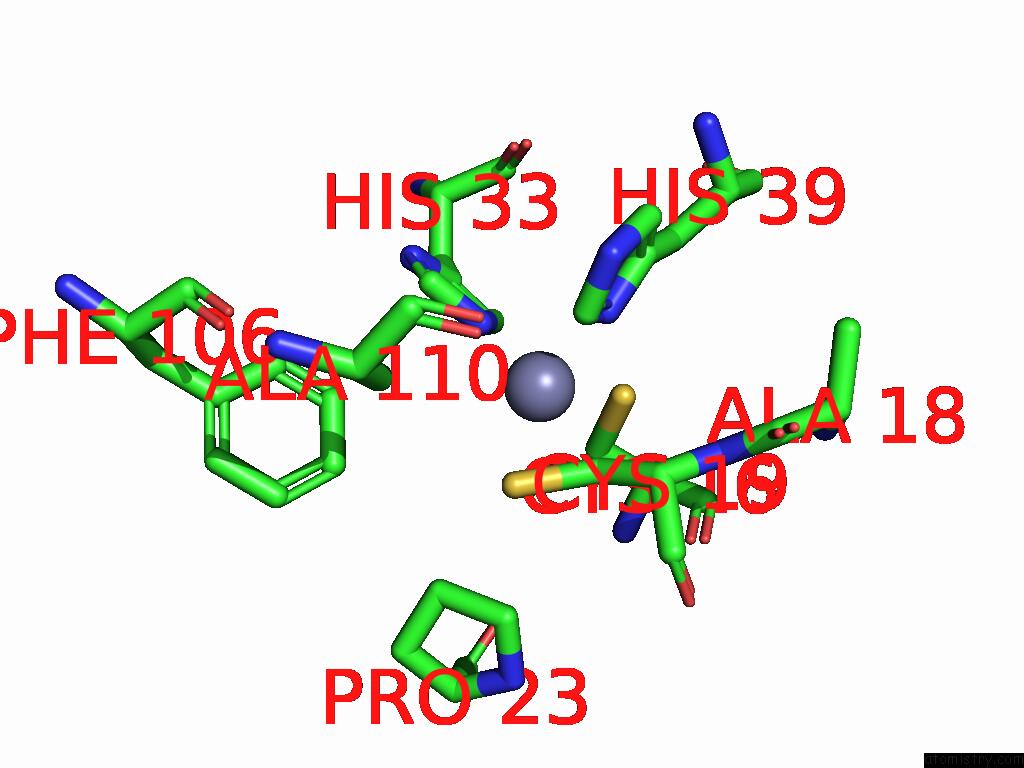
Mono view
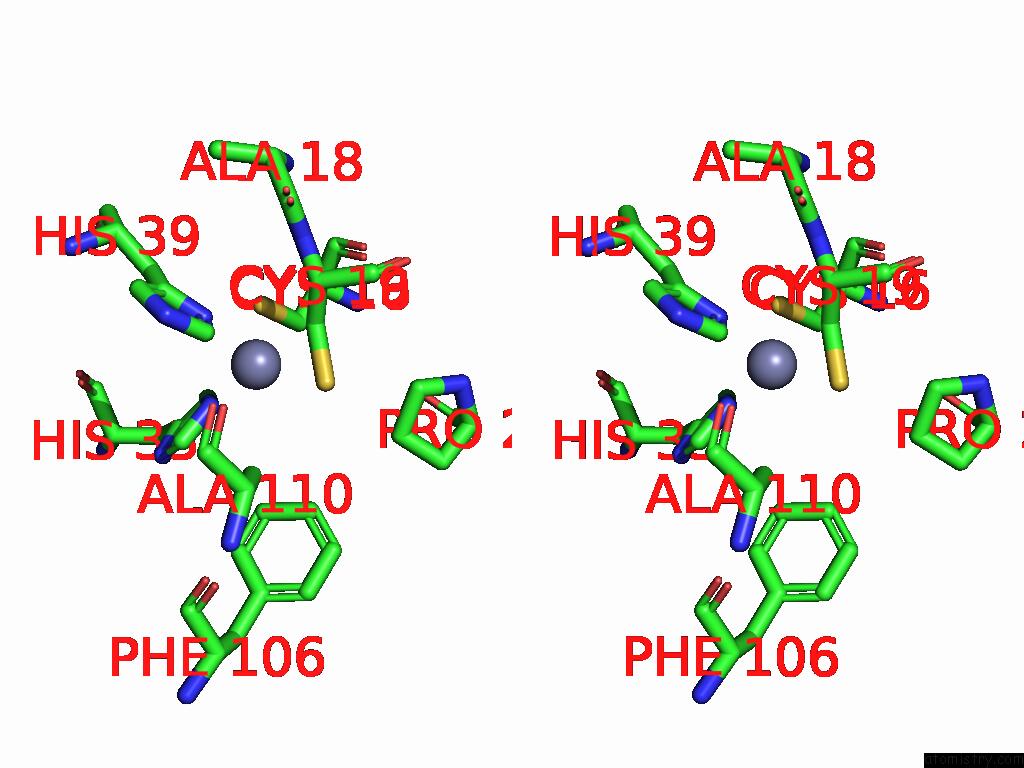
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 3 of Sars-Cov-2 Replication-Transcription Complex Has A Dimer Architecture (Local Drtc) in Post-Capping State within 5.0Å range:
|
Zinc binding site 4 out of 8 in 9imm
Go back to
Zinc binding site 4 out
of 8 in the Sars-Cov-2 Replication-Transcription Complex Has A Dimer Architecture (Local Drtc) in Post-Capping State
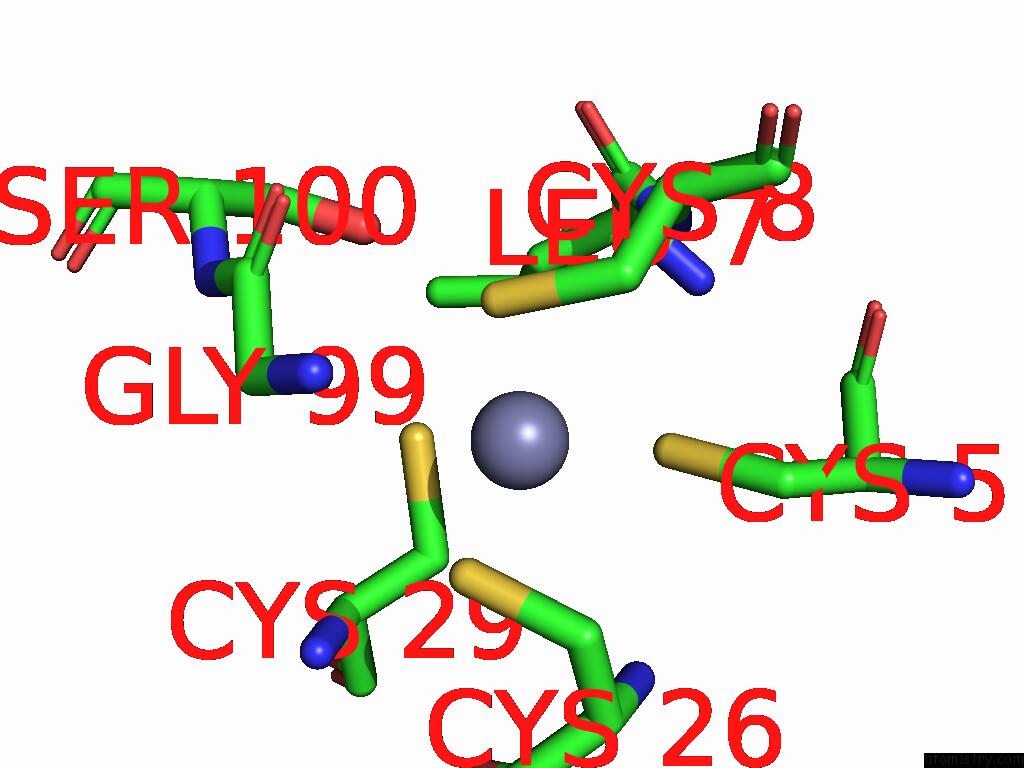
Mono view
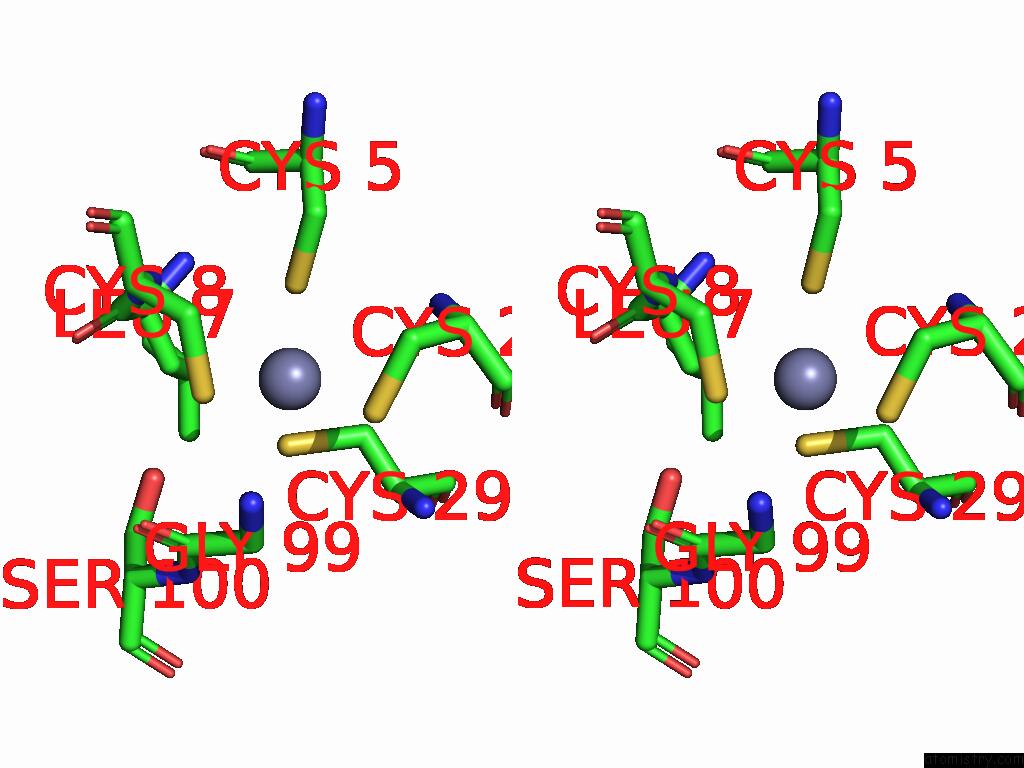
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 4 of Sars-Cov-2 Replication-Transcription Complex Has A Dimer Architecture (Local Drtc) in Post-Capping State within 5.0Å range:
|
Zinc binding site 5 out of 8 in 9imm
Go back to
Zinc binding site 5 out
of 8 in the Sars-Cov-2 Replication-Transcription Complex Has A Dimer Architecture (Local Drtc) in Post-Capping State
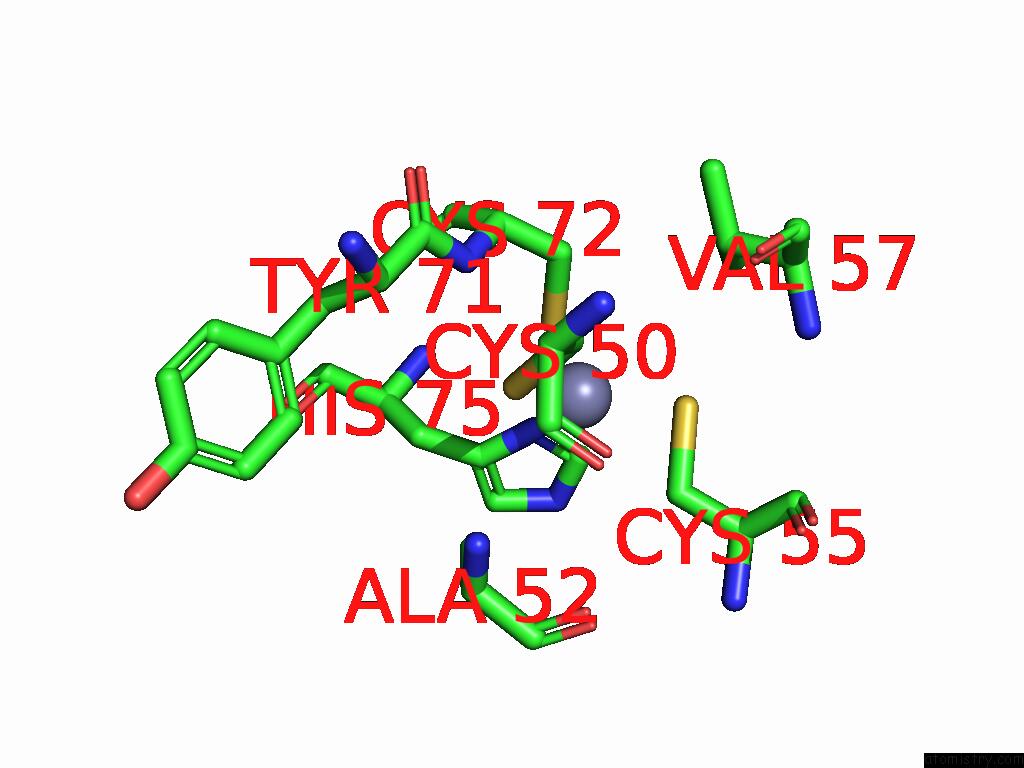
Mono view
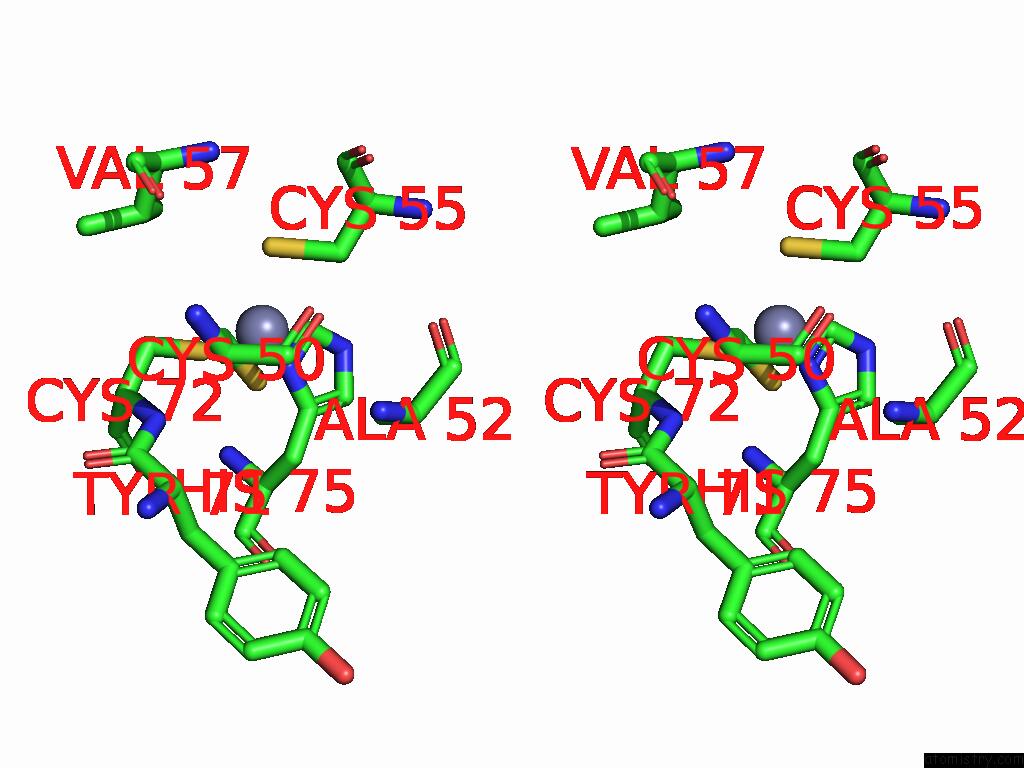
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 5 of Sars-Cov-2 Replication-Transcription Complex Has A Dimer Architecture (Local Drtc) in Post-Capping State within 5.0Å range:
|
Zinc binding site 6 out of 8 in 9imm
Go back to
Zinc binding site 6 out
of 8 in the Sars-Cov-2 Replication-Transcription Complex Has A Dimer Architecture (Local Drtc) in Post-Capping State
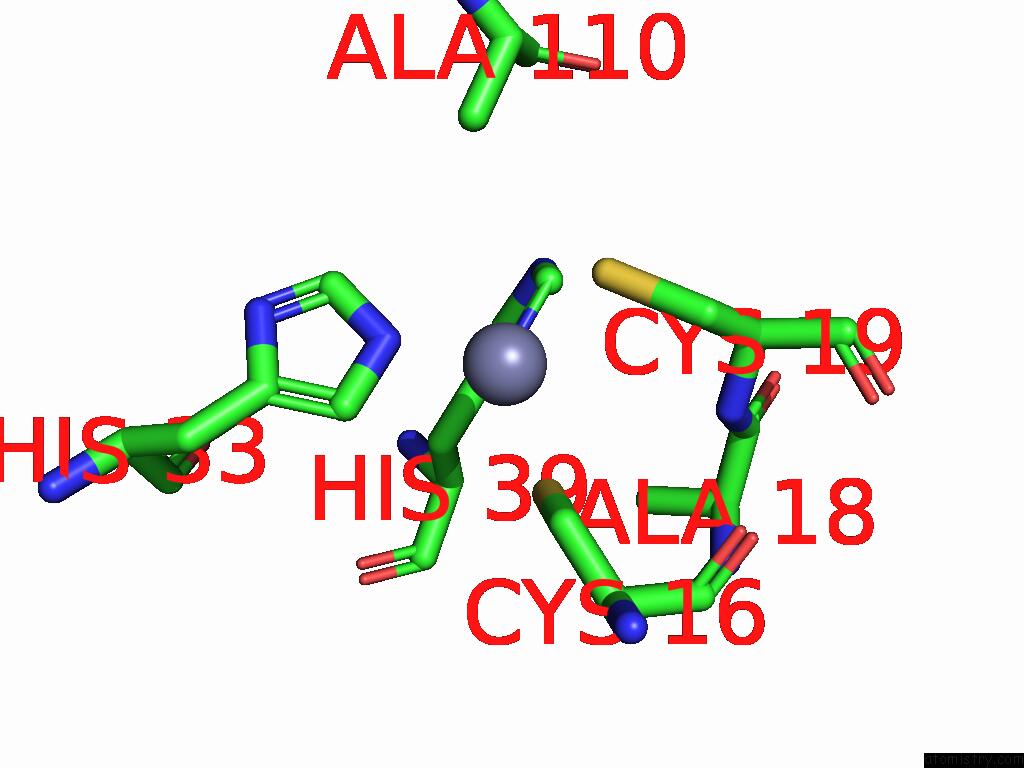
Mono view
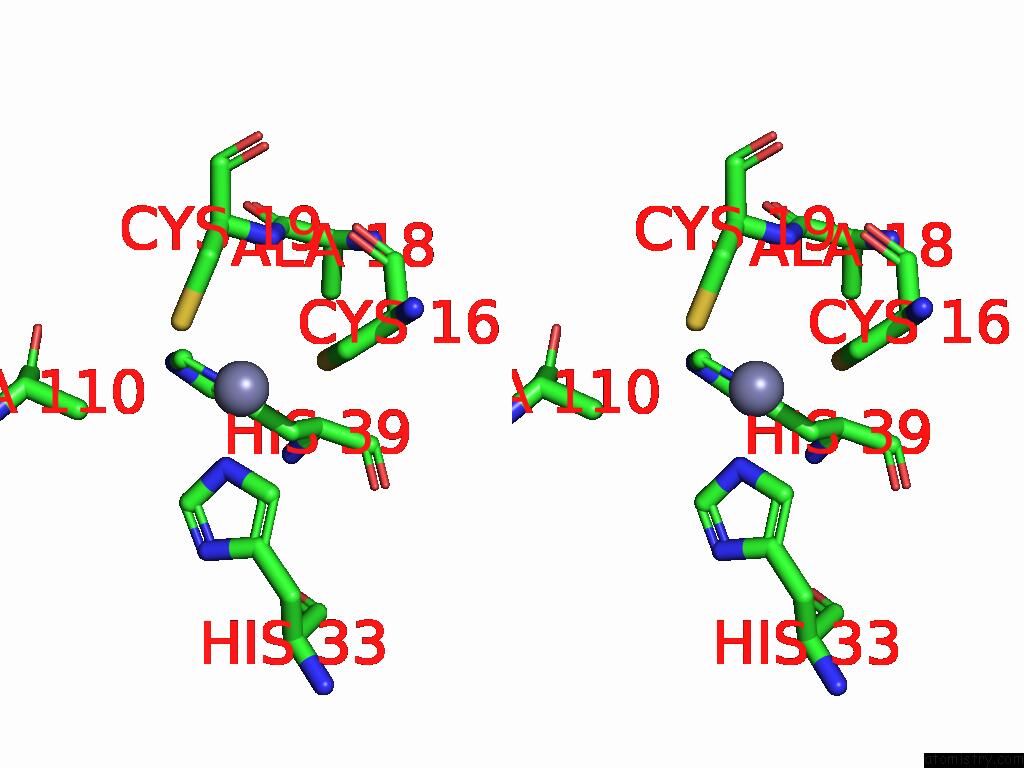
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 6 of Sars-Cov-2 Replication-Transcription Complex Has A Dimer Architecture (Local Drtc) in Post-Capping State within 5.0Å range:
|
Zinc binding site 7 out of 8 in 9imm
Go back to
Zinc binding site 7 out
of 8 in the Sars-Cov-2 Replication-Transcription Complex Has A Dimer Architecture (Local Drtc) in Post-Capping State
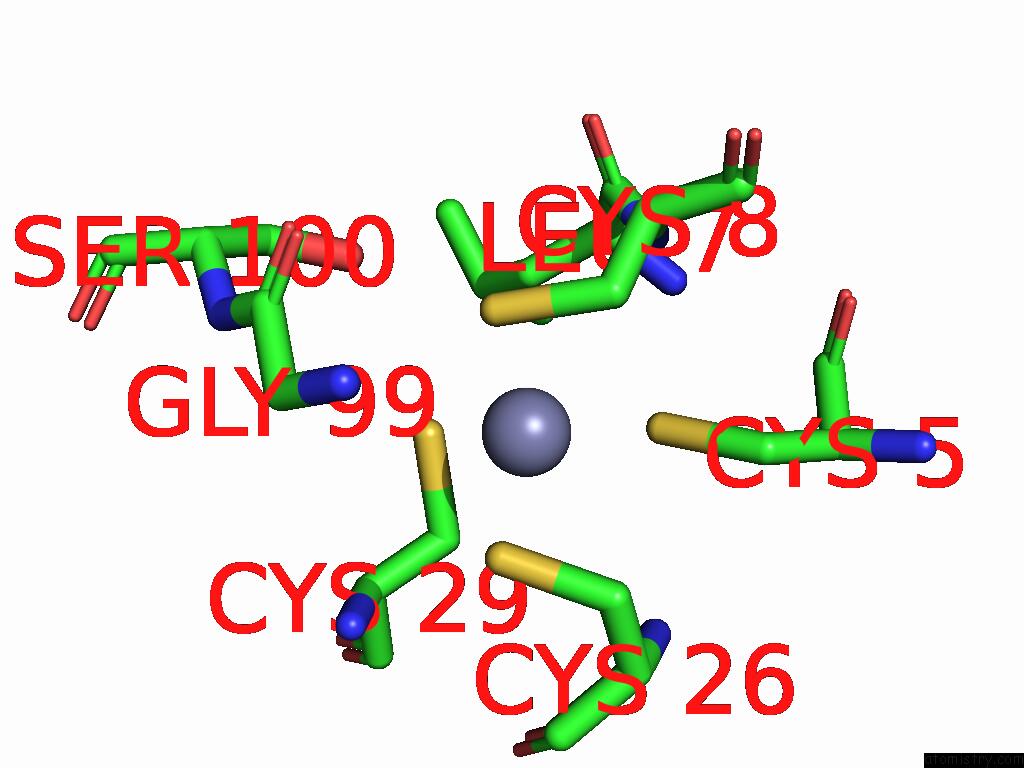
Mono view
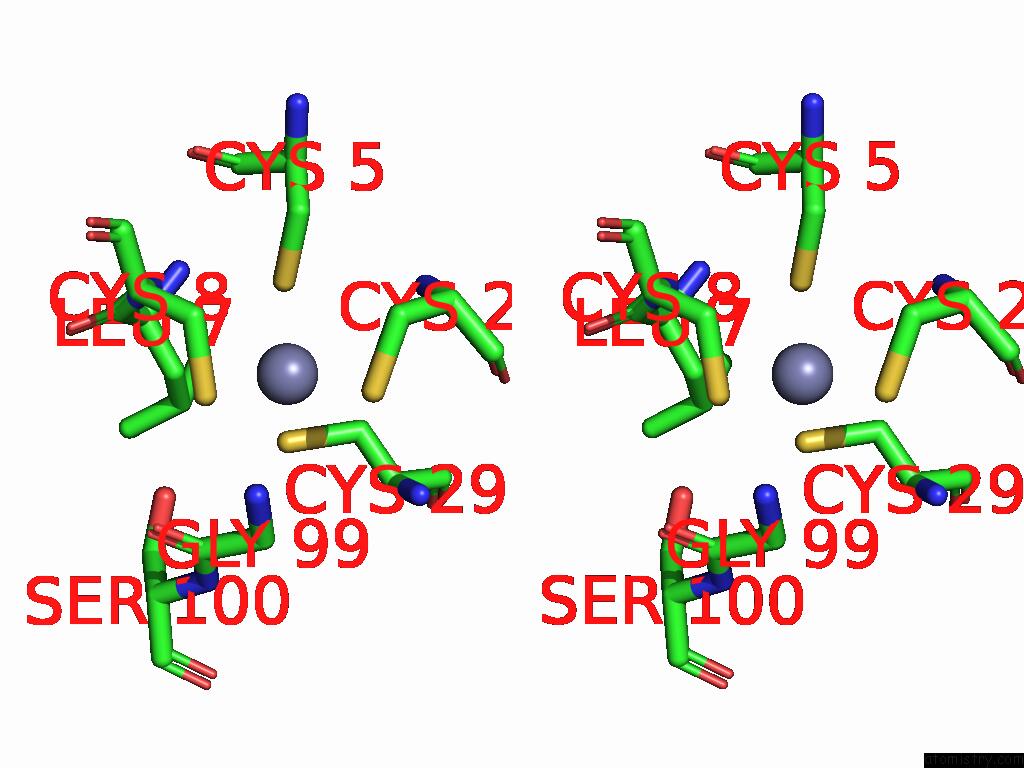
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 7 of Sars-Cov-2 Replication-Transcription Complex Has A Dimer Architecture (Local Drtc) in Post-Capping State within 5.0Å range:
|
Zinc binding site 8 out of 8 in 9imm
Go back to
Zinc binding site 8 out
of 8 in the Sars-Cov-2 Replication-Transcription Complex Has A Dimer Architecture (Local Drtc) in Post-Capping State
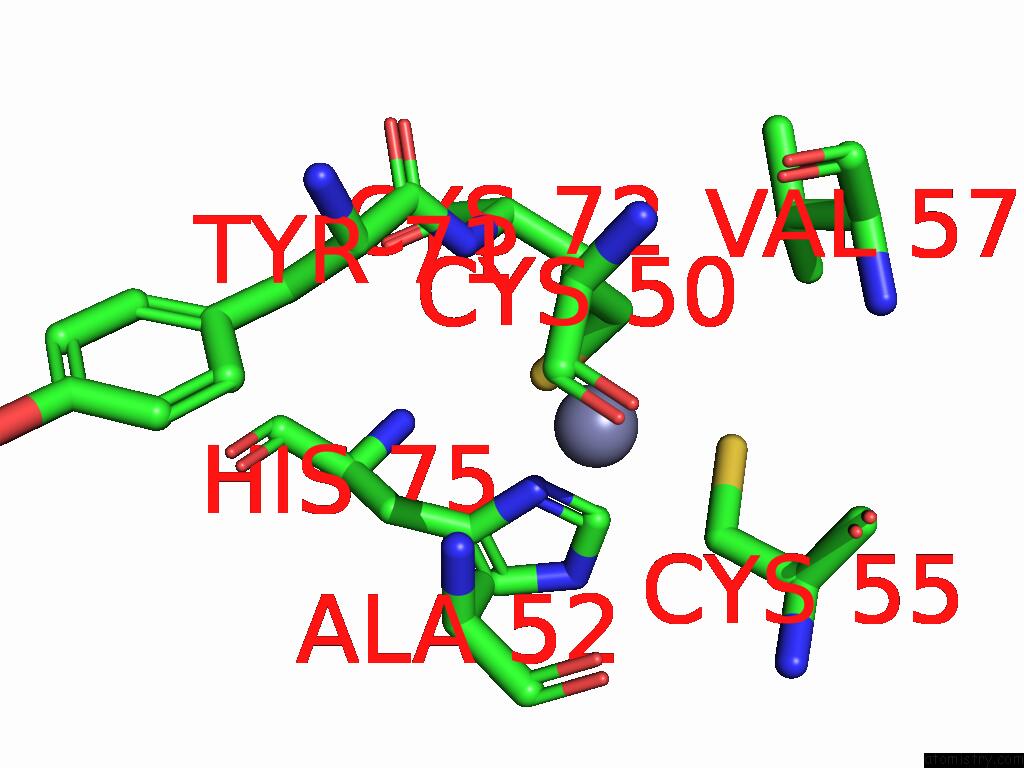
Mono view
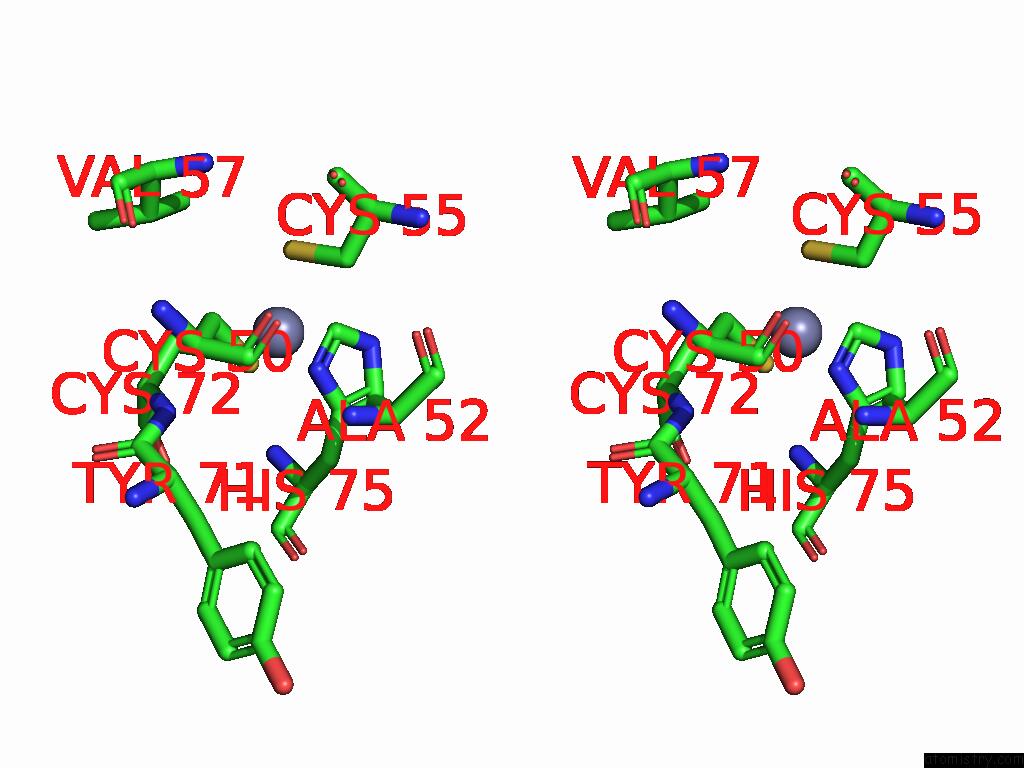
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 8 of Sars-Cov-2 Replication-Transcription Complex Has A Dimer Architecture (Local Drtc) in Post-Capping State within 5.0Å range:
|
Reference:
L.M.Yan,
Z.H.Rao,
Z.Y.Lou.
Structural Basis For the Concurrence of Template Recycling and Rna Capping in Sars-Cov-2 To Be Published.
Page generated: Tue Aug 26 22:02:43 2025
Last articles
Zn in 9QM9Zn in 9S44
Zn in 9OFE
Zn in 9OFC
Zn in 9OFD
Zn in 9OF1
Zn in 9OFB
Zn in 9N0J
Zn in 9M5X
Zn in 9LGI