Zinc »
PDB 8vne-8w34 »
8vrt »
Zinc in PDB 8vrt: The Structure of LSD1-Corest-HDAC1 in Complex with KBTBD4R313PRR Mutant
Enzymatic activity of The Structure of LSD1-Corest-HDAC1 in Complex with KBTBD4R313PRR Mutant
All present enzymatic activity of The Structure of LSD1-Corest-HDAC1 in Complex with KBTBD4R313PRR Mutant:
3.5.1.98;
3.5.1.98;
Zinc Binding Sites:
The binding sites of Zinc atom in the The Structure of LSD1-Corest-HDAC1 in Complex with KBTBD4R313PRR Mutant
(pdb code 8vrt). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total only one binding site of Zinc was determined in the The Structure of LSD1-Corest-HDAC1 in Complex with KBTBD4R313PRR Mutant, PDB code: 8vrt:
In total only one binding site of Zinc was determined in the The Structure of LSD1-Corest-HDAC1 in Complex with KBTBD4R313PRR Mutant, PDB code: 8vrt:
Zinc binding site 1 out of 1 in 8vrt
Go back to
Zinc binding site 1 out
of 1 in the The Structure of LSD1-Corest-HDAC1 in Complex with KBTBD4R313PRR Mutant
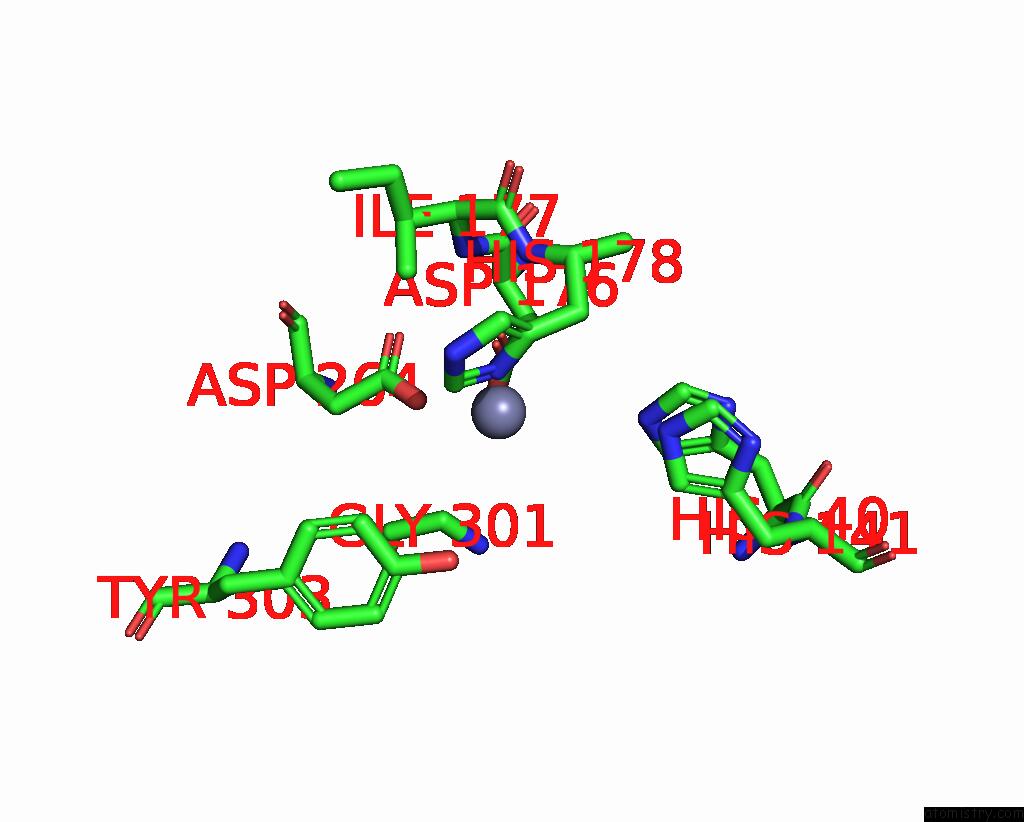
Mono view
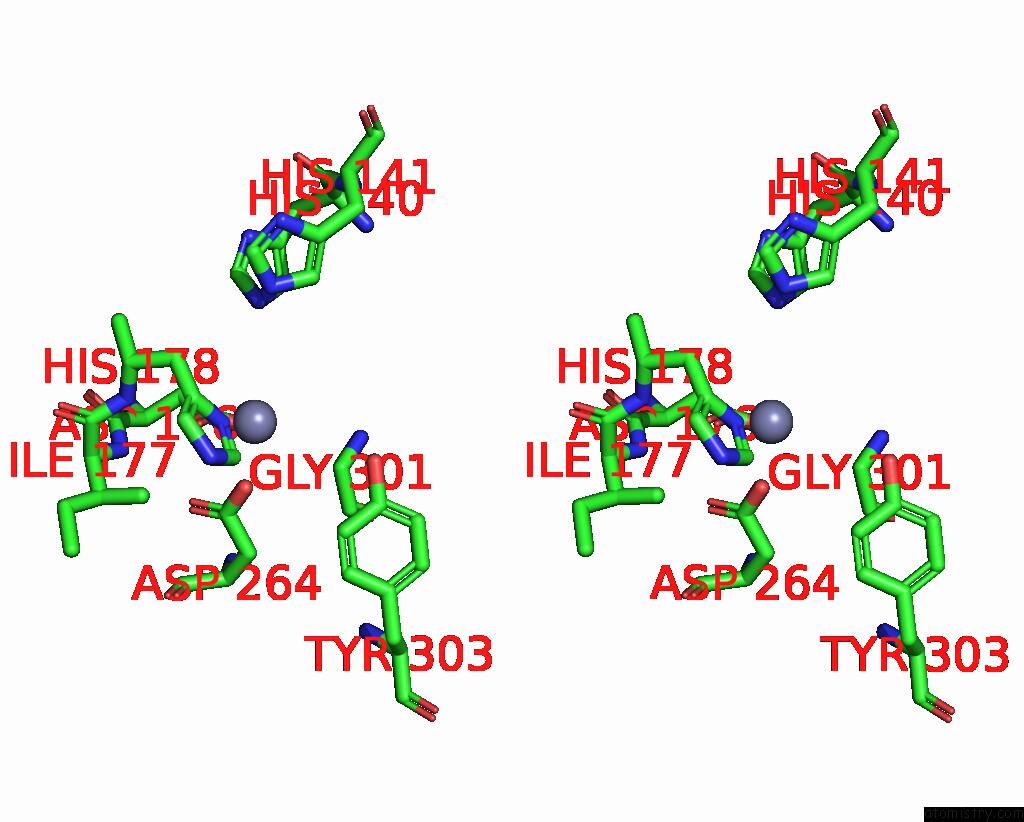
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of The Structure of LSD1-Corest-HDAC1 in Complex with KBTBD4R313PRR Mutant within 5.0Å range:
|
Reference:
X.Xie,
B.Liau,
N.Zheng.
Molecular Mechanism Underlying KBTBD4 Cancer Mutation-Driven Degradation of Hdac Corepressor Complexes To Be Published.
Page generated: Fri Aug 22 14:53:27 2025
Last articles
Mn in 9LJUMn in 9LJW
Mn in 9LJS
Mn in 9LJR
Mn in 9LJT
Mn in 9LJV
Mg in 9UA2
Mg in 9R96
Mg in 9VM1
Mg in 9P01