Zinc »
PDB 8rfj-8rsn »
8rhz »
Zinc in PDB 8rhz: Structure of CUL9-RBX1 Ubiquitin E3 Ligase Complex in Unneddylated Conformation - Symmetry Expanded Unneddylated Dimer
Enzymatic activity of Structure of CUL9-RBX1 Ubiquitin E3 Ligase Complex in Unneddylated Conformation - Symmetry Expanded Unneddylated Dimer
All present enzymatic activity of Structure of CUL9-RBX1 Ubiquitin E3 Ligase Complex in Unneddylated Conformation - Symmetry Expanded Unneddylated Dimer:
2.3.2.27; 2.3.2.32;
2.3.2.27; 2.3.2.32;
Zinc Binding Sites:
The binding sites of Zinc atom in the Structure of CUL9-RBX1 Ubiquitin E3 Ligase Complex in Unneddylated Conformation - Symmetry Expanded Unneddylated Dimer
(pdb code 8rhz). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 6 binding sites of Zinc where determined in the Structure of CUL9-RBX1 Ubiquitin E3 Ligase Complex in Unneddylated Conformation - Symmetry Expanded Unneddylated Dimer, PDB code: 8rhz:
Jump to Zinc binding site number: 1; 2; 3; 4; 5; 6;
In total 6 binding sites of Zinc where determined in the Structure of CUL9-RBX1 Ubiquitin E3 Ligase Complex in Unneddylated Conformation - Symmetry Expanded Unneddylated Dimer, PDB code: 8rhz:
Jump to Zinc binding site number: 1; 2; 3; 4; 5; 6;
Zinc binding site 1 out of 6 in 8rhz
Go back to
Zinc binding site 1 out
of 6 in the Structure of CUL9-RBX1 Ubiquitin E3 Ligase Complex in Unneddylated Conformation - Symmetry Expanded Unneddylated Dimer
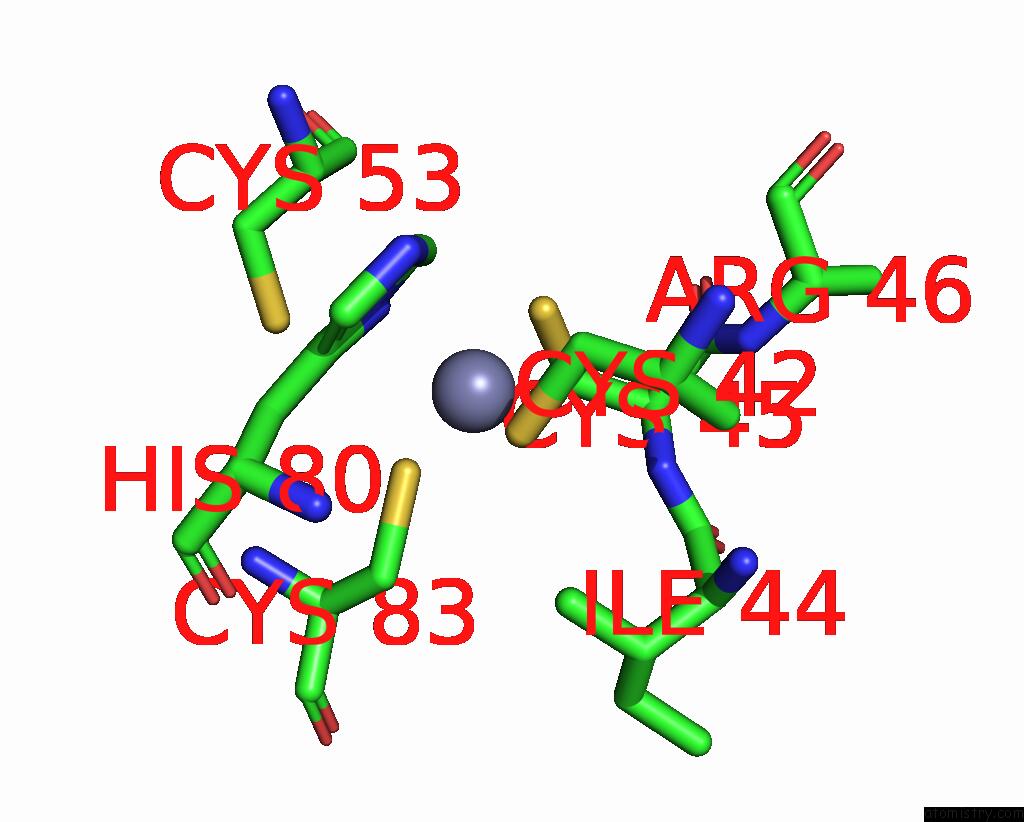
Mono view
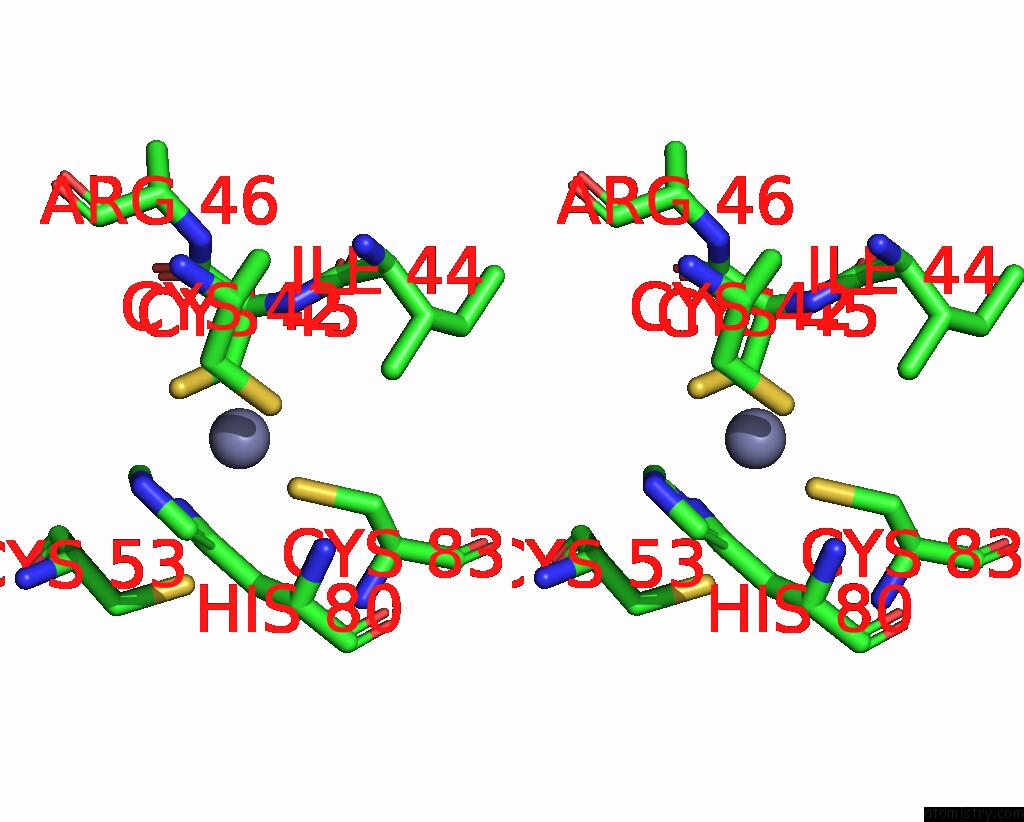
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Structure of CUL9-RBX1 Ubiquitin E3 Ligase Complex in Unneddylated Conformation - Symmetry Expanded Unneddylated Dimer within 5.0Å range:
|
Zinc binding site 2 out of 6 in 8rhz
Go back to
Zinc binding site 2 out
of 6 in the Structure of CUL9-RBX1 Ubiquitin E3 Ligase Complex in Unneddylated Conformation - Symmetry Expanded Unneddylated Dimer
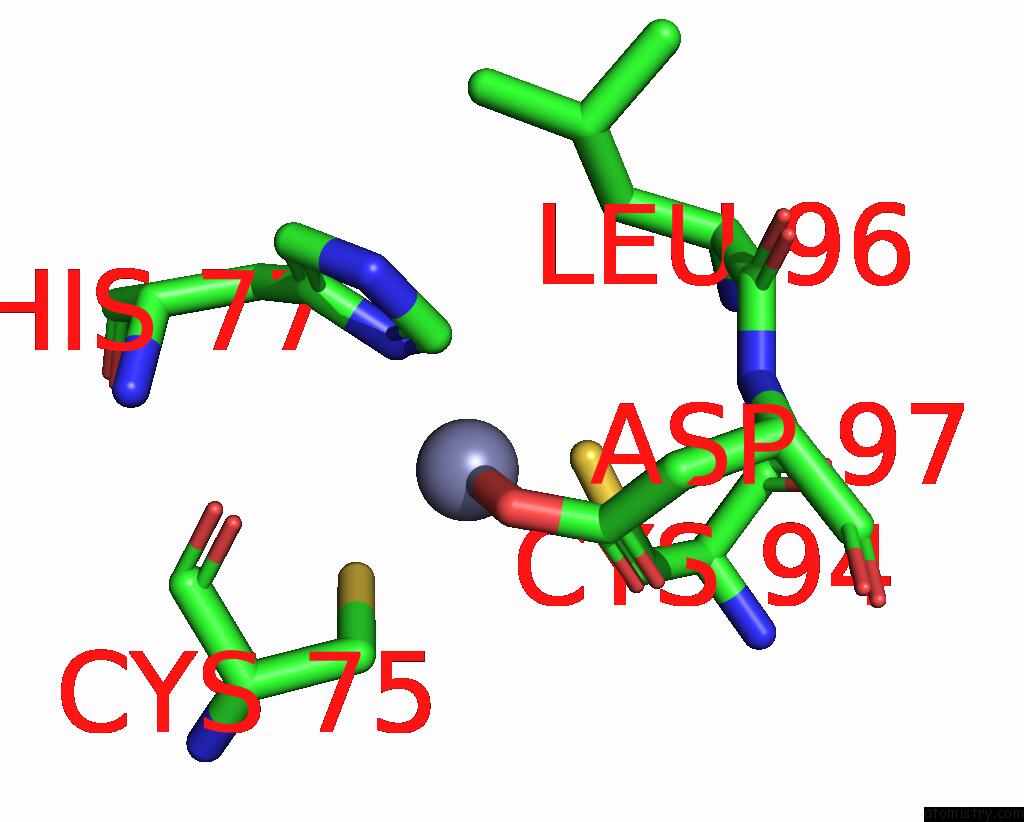
Mono view
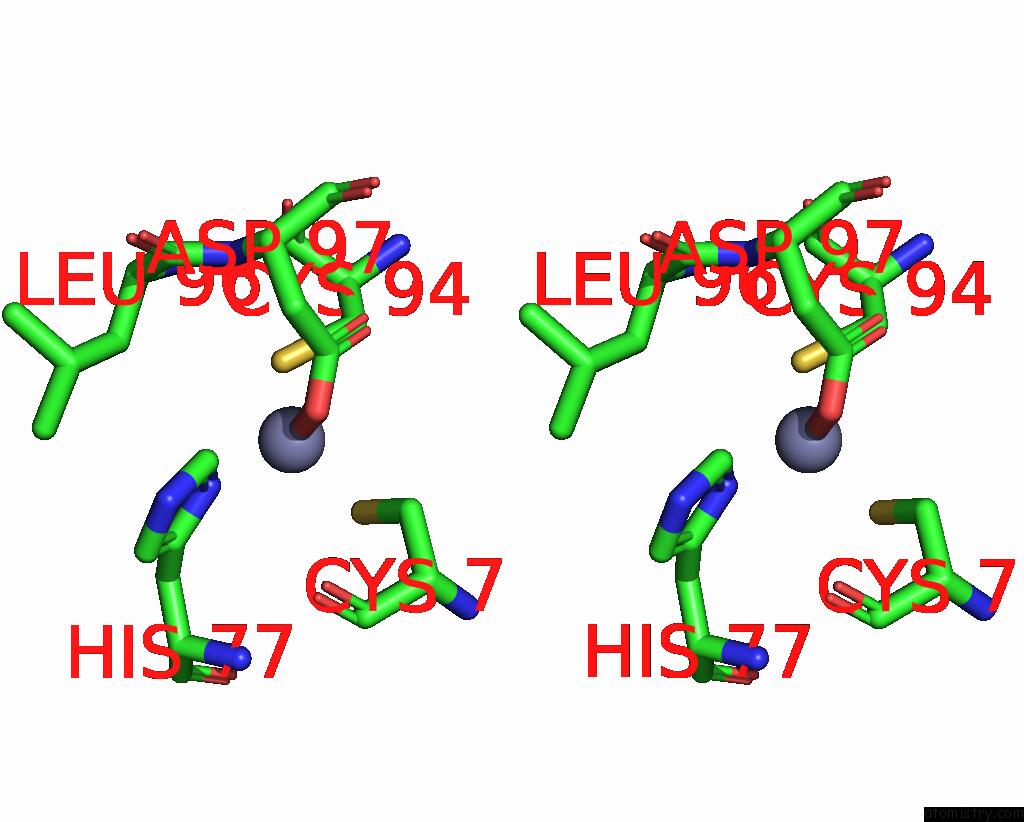
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of Structure of CUL9-RBX1 Ubiquitin E3 Ligase Complex in Unneddylated Conformation - Symmetry Expanded Unneddylated Dimer within 5.0Å range:
|
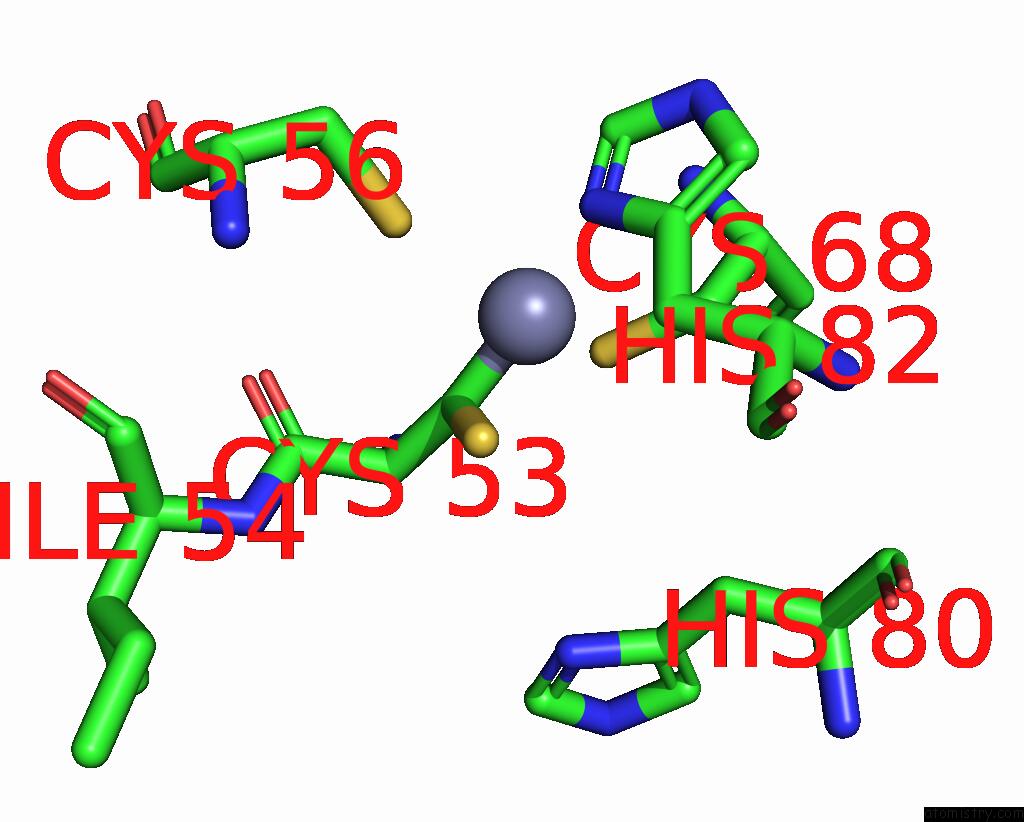
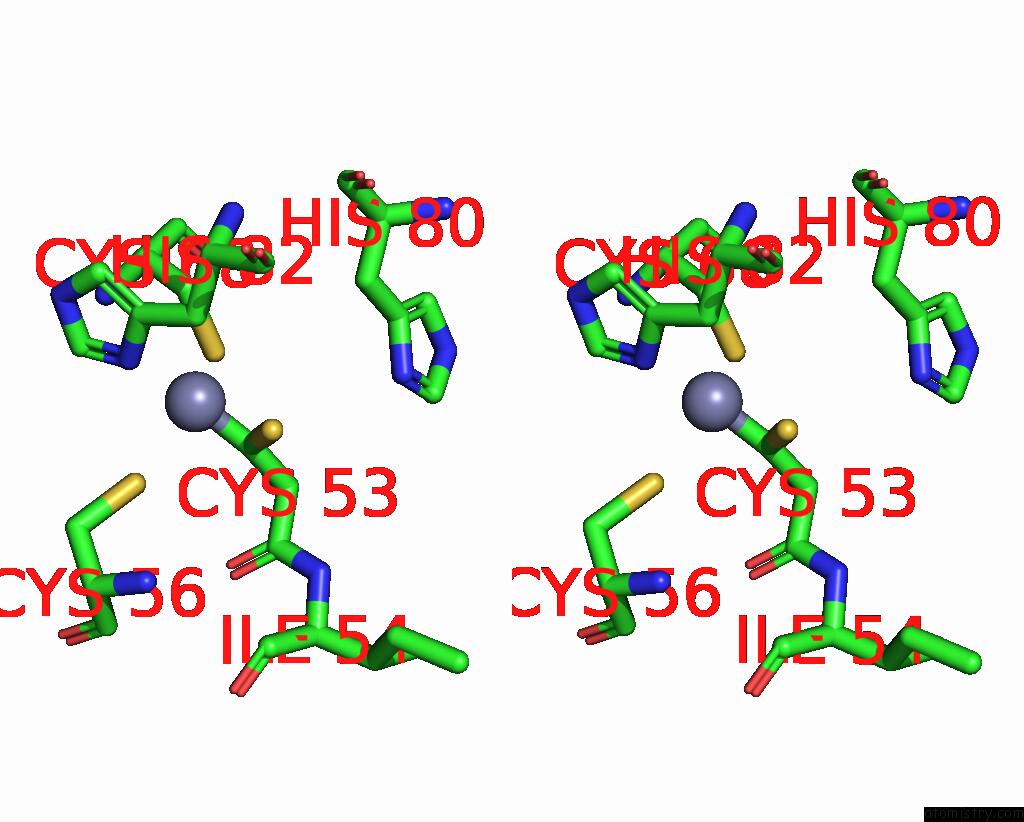
Zinc binding site 3 out of 6 in 8rhz
Go back to
Zinc binding site 3 out
of 6 in the Structure of CUL9-RBX1 Ubiquitin E3 Ligase Complex in Unneddylated Conformation - Symmetry Expanded Unneddylated Dimer
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 3 of Structure of CUL9-RBX1 Ubiquitin E3 Ligase Complex in Unneddylated Conformation - Symmetry Expanded Unneddylated Dimer within 5.0Å range:
|
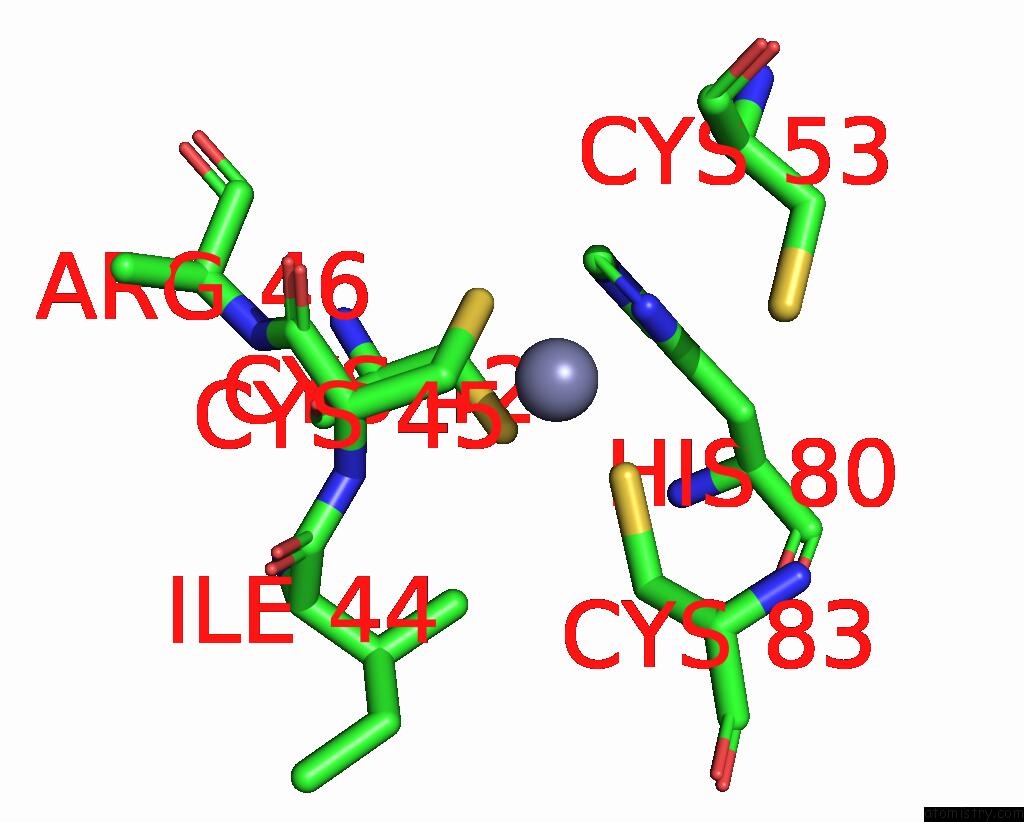
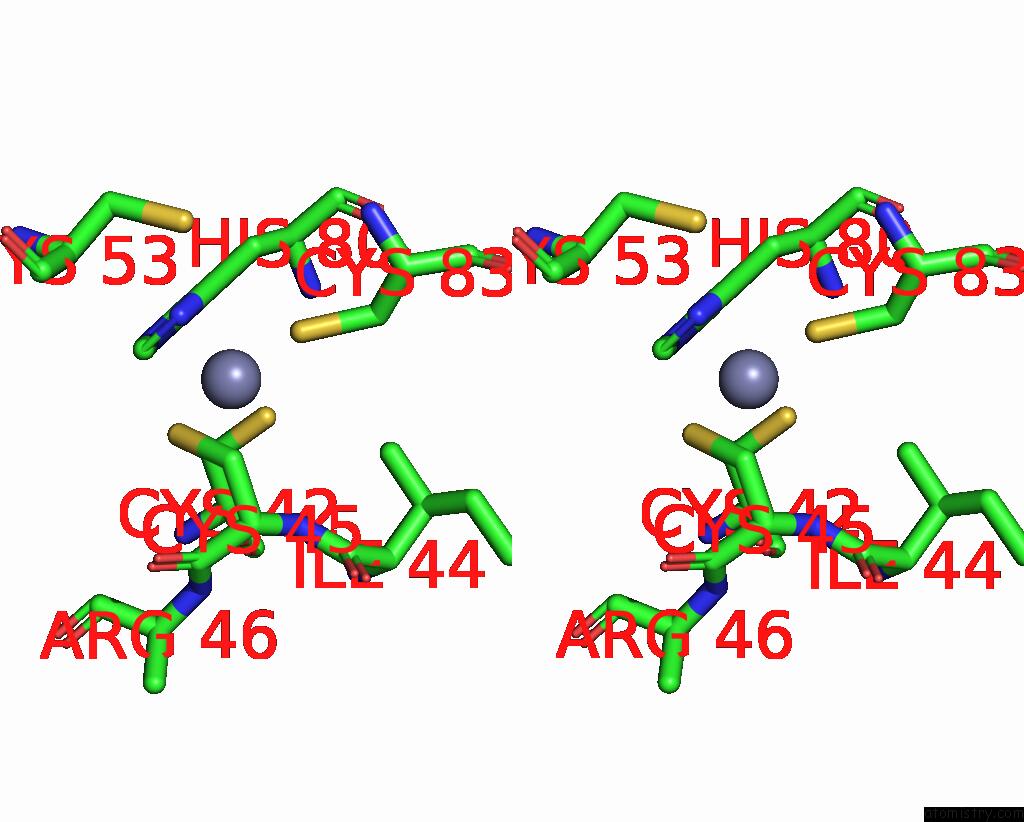
Zinc binding site 4 out of 6 in 8rhz
Go back to
Zinc binding site 4 out
of 6 in the Structure of CUL9-RBX1 Ubiquitin E3 Ligase Complex in Unneddylated Conformation - Symmetry Expanded Unneddylated Dimer
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 4 of Structure of CUL9-RBX1 Ubiquitin E3 Ligase Complex in Unneddylated Conformation - Symmetry Expanded Unneddylated Dimer within 5.0Å range:
|
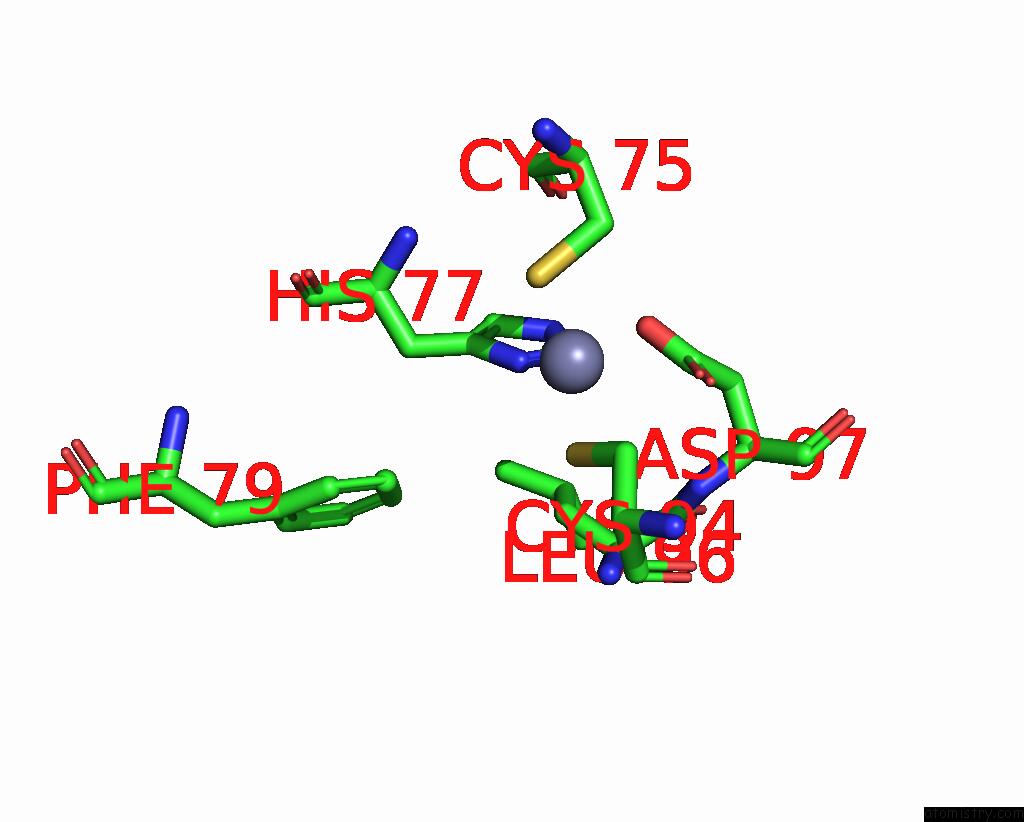
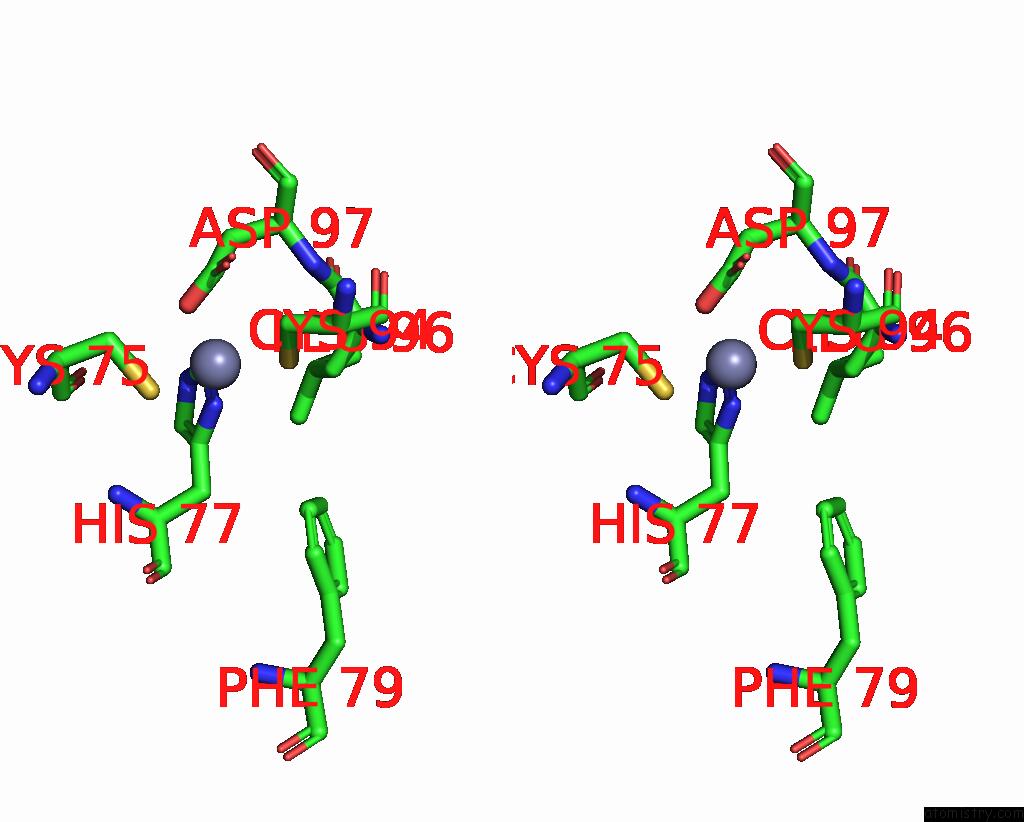
Zinc binding site 5 out of 6 in 8rhz
Go back to
Zinc binding site 5 out
of 6 in the Structure of CUL9-RBX1 Ubiquitin E3 Ligase Complex in Unneddylated Conformation - Symmetry Expanded Unneddylated Dimer
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 5 of Structure of CUL9-RBX1 Ubiquitin E3 Ligase Complex in Unneddylated Conformation - Symmetry Expanded Unneddylated Dimer within 5.0Å range:
|
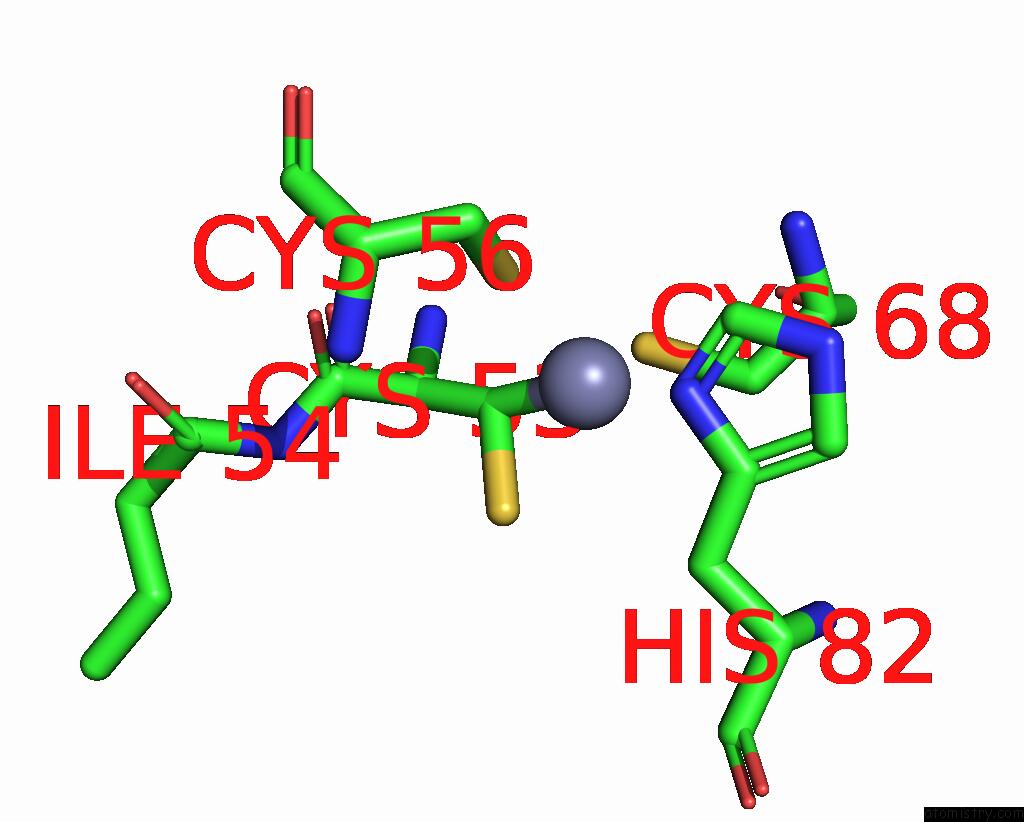
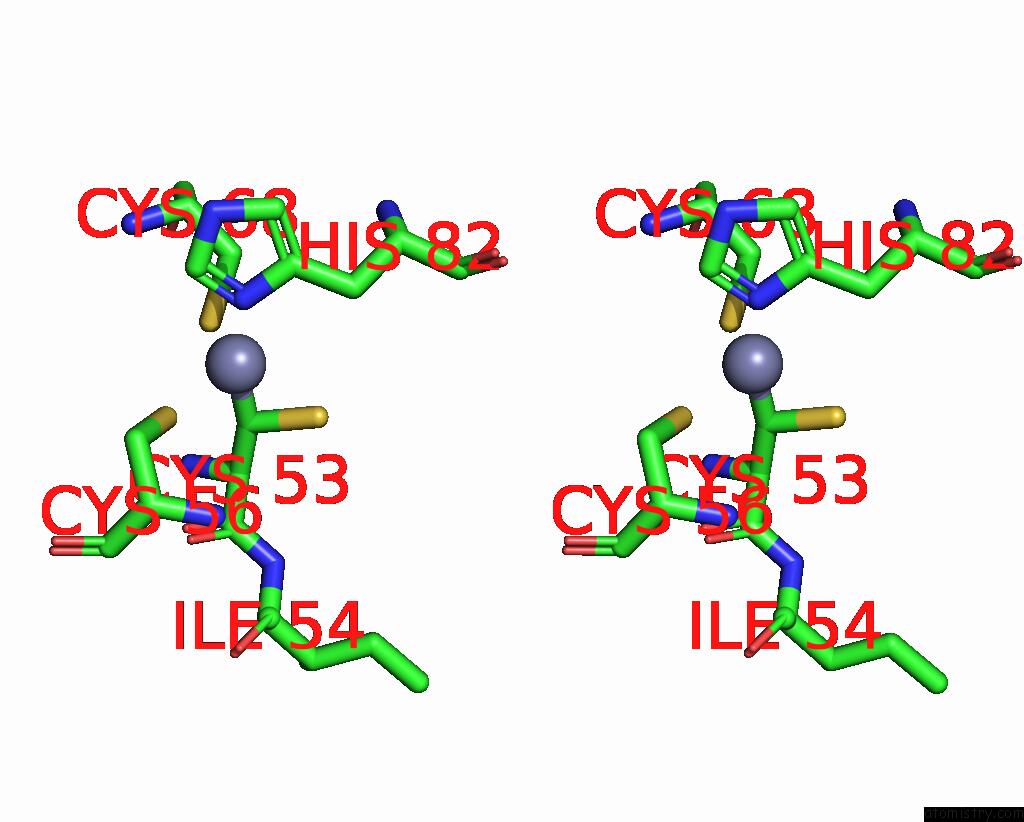
Zinc binding site 6 out of 6 in 8rhz
Go back to
Zinc binding site 6 out
of 6 in the Structure of CUL9-RBX1 Ubiquitin E3 Ligase Complex in Unneddylated Conformation - Symmetry Expanded Unneddylated Dimer
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 6 of Structure of CUL9-RBX1 Ubiquitin E3 Ligase Complex in Unneddylated Conformation - Symmetry Expanded Unneddylated Dimer within 5.0Å range:
|
Reference:
L.V.M.Hopf,
D.Horn-Ghetko,
B.A.Schulman.
Noncanonical Assembly, Neddylation and Chimeric Cullin Ring/Rbr Ubiquitylation By the 1.8 Mda CUL9 E3 Ligase Complex Nat.Struct.Mol.Biol. 2024.
ISSN: ESSN 1545-9985
DOI: 10.1038/S41594-024-01257-Y
Page generated: Fri Aug 22 13:01:01 2025
ISSN: ESSN 1545-9985
DOI: 10.1038/S41594-024-01257-Y
Last articles
Zn in 9CDHZn in 9CCH
Zn in 9CDG
Zn in 9CDF
Zn in 9CBT
Zn in 9C8U
Zn in 9CBR
Zn in 9CBP
Zn in 9CAH
Zn in 9CBQ